Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
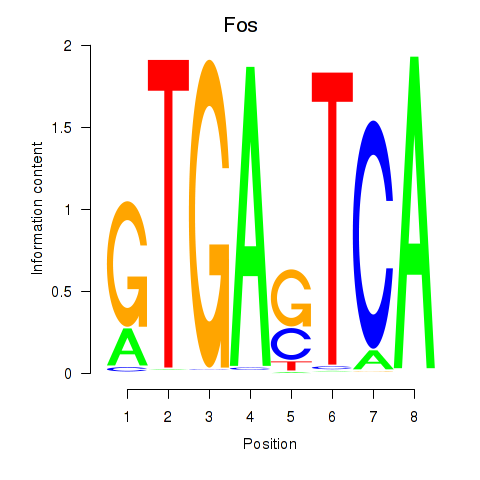
Results for Fos
Z-value: 2.03
Transcription factors associated with Fos
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fos
|
ENSMUSG00000021250.14 | FBJ osteosarcoma oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fos | mm39_v1_chr12_+_85520652_85520672 | 0.63 | 3.9e-05 | Click! |
Activity profile of Fos motif
Sorted Z-values of Fos motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102255999 | 34.45 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr7_-_45570254 | 28.21 |
ENSMUST00000164119.3
|
Emp3
|
epithelial membrane protein 3 |
| chr7_-_45570538 | 27.83 |
ENSMUST00000210297.2
|
Emp3
|
epithelial membrane protein 3 |
| chr1_-_132318039 | 26.00 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr7_+_24069680 | 22.80 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr3_-_88410495 | 20.73 |
ENSMUST00000120377.8
ENSMUST00000029699.13 |
Lmna
|
lamin A |
| chr7_-_45570674 | 16.34 |
ENSMUST00000210939.2
|
Emp3
|
epithelial membrane protein 3 |
| chr2_+_164790139 | 16.02 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr7_+_78563964 | 15.90 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr2_+_13578738 | 15.59 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr8_+_95710977 | 15.49 |
ENSMUST00000093271.8
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr7_+_78563513 | 15.20 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr7_-_126398343 | 14.23 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr2_+_84564394 | 14.09 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr10_+_128745214 | 13.84 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr7_-_125968653 | 13.78 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
| chr7_-_126398165 | 13.57 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chrX_-_7537580 | 13.20 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr1_+_174000304 | 13.12 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr7_-_143056252 | 12.91 |
ENSMUST00000010904.5
|
Phlda2
|
pleckstrin homology like domain, family A, member 2 |
| chr7_+_78564062 | 12.41 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chrX_-_142610371 | 12.14 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chr13_+_95833359 | 11.23 |
ENSMUST00000022182.5
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr7_+_110371811 | 11.21 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr9_+_51124983 | 10.77 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chrX_-_133483849 | 10.70 |
ENSMUST00000113213.2
ENSMUST00000033617.13 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr2_+_24226857 | 10.15 |
ENSMUST00000114487.9
ENSMUST00000142093.7 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr4_-_43558386 | 9.87 |
ENSMUST00000130353.2
|
Tln1
|
talin 1 |
| chr14_-_54949596 | 9.64 |
ENSMUST00000064290.8
|
Cebpe
|
CCAAT/enhancer binding protein (C/EBP), epsilon |
| chr4_-_43040278 | 9.58 |
ENSMUST00000107958.8
ENSMUST00000107959.8 ENSMUST00000152846.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr4_-_140501507 | 9.53 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr9_+_69360902 | 9.44 |
ENSMUST00000034756.15
ENSMUST00000123470.8 |
Anxa2
|
annexin A2 |
| chr7_+_18817767 | 9.32 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr12_+_103354915 | 9.26 |
ENSMUST00000101094.9
ENSMUST00000021620.13 |
Otub2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr6_+_135339543 | 9.08 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr17_+_35268942 | 8.96 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr9_+_69361348 | 8.72 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr8_+_94954280 | 8.40 |
ENSMUST00000109547.2
|
Nup93
|
nucleoporin 93 |
| chr3_+_95836558 | 7.96 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_+_172328768 | 7.85 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr17_+_46471950 | 7.50 |
ENSMUST00000024748.14
ENSMUST00000172170.8 |
Gtpbp2
|
GTP binding protein 2 |
| chr8_+_123008855 | 7.49 |
ENSMUST00000054052.15
|
Zfpm1
|
zinc finger protein, multitype 1 |
| chr11_+_116422712 | 7.37 |
ENSMUST00000100201.10
|
Sphk1
|
sphingosine kinase 1 |
| chr11_+_116422570 | 7.36 |
ENSMUST00000106387.9
|
Sphk1
|
sphingosine kinase 1 |
| chr10_+_111309020 | 7.25 |
ENSMUST00000065917.16
ENSMUST00000219961.2 ENSMUST00000217908.2 |
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr3_+_95836637 | 7.19 |
ENSMUST00000171368.8
ENSMUST00000168106.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr10_+_111309127 | 6.98 |
ENSMUST00000219143.2
|
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chrX_-_47543029 | 6.98 |
ENSMUST00000114958.8
|
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr11_+_116424082 | 6.94 |
ENSMUST00000154034.8
|
Sphk1
|
sphingosine kinase 1 |
| chr1_-_59012579 | 6.67 |
ENSMUST00000173590.2
ENSMUST00000027186.12 |
Trak2
|
trafficking protein, kinesin binding 2 |
| chr14_+_79753055 | 6.65 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chrX_-_165992311 | 6.51 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr6_+_135339929 | 6.43 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr3_+_95139598 | 6.28 |
ENSMUST00000107200.8
ENSMUST00000107199.2 |
Cdc42se1
|
CDC42 small effector 1 |
| chr5_+_35156389 | 6.27 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr14_-_32907023 | 6.20 |
ENSMUST00000130509.10
ENSMUST00000061753.15 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr10_+_79762858 | 6.19 |
ENSMUST00000019708.12
ENSMUST00000105377.8 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr8_+_95711037 | 6.08 |
ENSMUST00000211944.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chrX_-_165992145 | 6.04 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr6_+_42326760 | 6.02 |
ENSMUST00000203652.3
ENSMUST00000070635.13 |
Zyx
|
zyxin |
| chr6_+_42326934 | 5.96 |
ENSMUST00000203401.3
ENSMUST00000164375.4 |
Zyx
|
zyxin |
| chr7_-_19005721 | 5.92 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr6_-_113411718 | 5.86 |
ENSMUST00000113091.8
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr17_+_12338161 | 5.84 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr6_+_42326714 | 5.78 |
ENSMUST00000203846.3
|
Zyx
|
zyxin |
| chr5_+_100993996 | 5.72 |
ENSMUST00000112887.8
ENSMUST00000031255.15 |
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr11_-_53371050 | 5.70 |
ENSMUST00000104955.4
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr5_+_25452342 | 5.70 |
ENSMUST00000114950.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr17_-_57501170 | 5.70 |
ENSMUST00000005976.8
|
Tnfsf14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr17_+_29170924 | 5.68 |
ENSMUST00000153462.8
|
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr7_+_27186335 | 5.61 |
ENSMUST00000008528.8
|
Sertad1
|
SERTA domain containing 1 |
| chr15_-_64184485 | 5.60 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr18_+_82928959 | 5.58 |
ENSMUST00000171238.8
|
Zfp516
|
zinc finger protein 516 |
| chr10_+_80765900 | 5.49 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr11_+_78079243 | 5.31 |
ENSMUST00000002128.14
ENSMUST00000150941.8 |
Rab34
|
RAB34, member RAS oncogene family |
| chr5_-_134975773 | 5.28 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr17_+_29171655 | 5.27 |
ENSMUST00000117672.9
ENSMUST00000153831.9 |
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr2_-_113883285 | 5.26 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr2_+_30331839 | 5.20 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr11_+_61575245 | 5.03 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G |
| chr8_+_89015705 | 4.95 |
ENSMUST00000171456.9
|
Adcy7
|
adenylate cyclase 7 |
| chr17_-_7652863 | 4.90 |
ENSMUST00000070059.5
|
Unc93a2
|
unc-93 homolog A2 |
| chr3_-_10273628 | 4.87 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr10_+_80100812 | 4.84 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr11_+_78079631 | 4.79 |
ENSMUST00000056241.12
ENSMUST00000207728.2 |
Rab34
|
RAB34, member RAS oncogene family |
| chr4_+_132857816 | 4.52 |
ENSMUST00000084241.12
ENSMUST00000138831.2 |
Wasf2
|
WASP family, member 2 |
| chr5_+_100994230 | 4.48 |
ENSMUST00000092990.4
ENSMUST00000145612.2 |
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr4_-_6454262 | 4.36 |
ENSMUST00000029910.12
|
Nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr14_-_70415117 | 4.21 |
ENSMUST00000022681.11
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr7_-_30741532 | 4.21 |
ENSMUST00000160689.8
ENSMUST00000202395.4 ENSMUST00000162733.8 ENSMUST00000162087.8 ENSMUST00000009831.14 |
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr2_-_25086810 | 4.21 |
ENSMUST00000081869.7
|
Tor4a
|
torsin family 4, member A |
| chr16_+_35590745 | 4.18 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr12_+_111505216 | 4.14 |
ENSMUST00000050993.11
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr12_+_111505253 | 4.10 |
ENSMUST00000220803.2
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr11_-_78950641 | 4.06 |
ENSMUST00000226282.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr18_-_38131766 | 4.05 |
ENSMUST00000236588.2
ENSMUST00000237272.2 ENSMUST00000236134.2 |
Arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr3_+_103009920 | 4.04 |
ENSMUST00000173206.8
|
Dennd2c
|
DENN/MADD domain containing 2C |
| chr15_-_90563510 | 3.98 |
ENSMUST00000014777.9
ENSMUST00000064391.12 |
Cpne8
|
copine VIII |
| chr4_+_43506966 | 3.91 |
ENSMUST00000030183.10
|
Car9
|
carbonic anhydrase 9 |
| chr7_+_126376353 | 3.90 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr7_-_105131407 | 3.86 |
ENSMUST00000047040.4
|
Cavin3
|
caveolae associated 3 |
| chr6_+_42326980 | 3.85 |
ENSMUST00000203849.2
|
Zyx
|
zyxin |
| chr3_+_14706781 | 3.77 |
ENSMUST00000029071.9
|
Car13
|
carbonic anhydrase 13 |
| chr12_+_31315270 | 3.73 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chrX_-_100266032 | 3.71 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr15_+_84553801 | 3.66 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chrX_+_95498965 | 3.61 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr5_+_35156454 | 3.53 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr7_-_30741497 | 3.52 |
ENSMUST00000162116.8
ENSMUST00000159924.8 |
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr9_-_78388080 | 3.50 |
ENSMUST00000156988.2
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr8_-_25581354 | 3.49 |
ENSMUST00000125466.2
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chrX_+_158623460 | 3.38 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr6_+_42326528 | 3.28 |
ENSMUST00000203329.3
|
Zyx
|
zyxin |
| chr19_-_34143437 | 3.28 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr7_+_126184108 | 3.28 |
ENSMUST00000039522.8
|
Apobr
|
apolipoprotein B receptor |
| chr4_-_3835595 | 3.27 |
ENSMUST00000138502.2
|
Rps20
|
ribosomal protein S20 |
| chr1_-_52230062 | 3.19 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr2_-_101627999 | 3.16 |
ENSMUST00000171088.8
ENSMUST00000043845.14 |
Prr5l
|
proline rich 5 like |
| chr10_+_97318223 | 3.11 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr10_-_76562002 | 3.11 |
ENSMUST00000001147.5
|
Col6a1
|
collagen, type VI, alpha 1 |
| chr13_-_21964747 | 3.03 |
ENSMUST00000080511.3
|
H1f5
|
H1.5 linker histone, cluster member |
| chr3_-_122828592 | 3.00 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr2_-_25086770 | 2.99 |
ENSMUST00000142857.2
ENSMUST00000137920.2 |
Tor4a
|
torsin family 4, member A |
| chr1_-_153061758 | 2.96 |
ENSMUST00000185356.7
|
Lamc2
|
laminin, gamma 2 |
| chr1_+_107439145 | 2.95 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr11_-_76400245 | 2.95 |
ENSMUST00000094012.11
|
Abr
|
active BCR-related gene |
| chr2_-_25114660 | 2.93 |
ENSMUST00000043584.5
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr11_+_78079562 | 2.92 |
ENSMUST00000108322.9
|
Rab34
|
RAB34, member RAS oncogene family |
| chr10_+_79763164 | 2.92 |
ENSMUST00000105376.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr2_-_44817173 | 2.92 |
ENSMUST00000130991.8
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr1_+_40554513 | 2.90 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr7_-_30741237 | 2.90 |
ENSMUST00000161805.8
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr10_+_94386714 | 2.89 |
ENSMUST00000148910.3
ENSMUST00000117460.8 |
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr14_+_25459690 | 2.86 |
ENSMUST00000007961.15
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr7_-_101899294 | 2.80 |
ENSMUST00000106923.2
ENSMUST00000098230.11 |
Rhog
|
ras homolog family member G |
| chr17_+_36152383 | 2.76 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr17_-_48145466 | 2.76 |
ENSMUST00000066368.13
|
Mdfi
|
MyoD family inhibitor |
| chr9_+_107879700 | 2.71 |
ENSMUST00000035214.11
ENSMUST00000176854.7 ENSMUST00000175874.2 |
Ip6k1
|
inositol hexaphosphate kinase 1 |
| chr10_+_128247598 | 2.71 |
ENSMUST00000096386.13
|
Rnf41
|
ring finger protein 41 |
| chr12_+_31315227 | 2.69 |
ENSMUST00000169088.8
|
Lamb1
|
laminin B1 |
| chr11_-_100098333 | 2.67 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr19_-_32188413 | 2.67 |
ENSMUST00000151289.9
|
Sgms1
|
sphingomyelin synthase 1 |
| chr18_-_43610829 | 2.67 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr17_-_45883421 | 2.67 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr9_-_71803354 | 2.62 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr18_+_23885390 | 2.60 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_+_128247492 | 2.60 |
ENSMUST00000171342.3
|
Rnf41
|
ring finger protein 41 |
| chr7_+_30450896 | 2.59 |
ENSMUST00000182229.8
ENSMUST00000080518.14 ENSMUST00000182227.8 ENSMUST00000182721.8 |
Sbsn
|
suprabasin |
| chr13_-_99027482 | 2.58 |
ENSMUST00000179301.8
ENSMUST00000179271.2 |
Tnpo1
|
transportin 1 |
| chr13_-_63006176 | 2.58 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr10_+_127126643 | 2.57 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr9_-_116004386 | 2.56 |
ENSMUST00000035014.8
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr11_+_20493306 | 2.54 |
ENSMUST00000093292.11
|
Sertad2
|
SERTA domain containing 2 |
| chr3_-_95139472 | 2.54 |
ENSMUST00000196025.5
ENSMUST00000198948.5 |
Mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr19_+_29229147 | 2.51 |
ENSMUST00000025705.7
ENSMUST00000065796.10 ENSMUST00000236990.2 |
Jak2
|
Janus kinase 2 |
| chrX_-_103714653 | 2.50 |
ENSMUST00000042070.6
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr2_+_155593030 | 2.49 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr4_-_6454097 | 2.48 |
ENSMUST00000124344.2
|
Nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr3_+_88528743 | 2.47 |
ENSMUST00000170653.9
ENSMUST00000177303.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr5_+_105563605 | 2.45 |
ENSMUST00000112707.3
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chrX_+_71006577 | 2.45 |
ENSMUST00000048790.7
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr16_-_44379226 | 2.45 |
ENSMUST00000114634.3
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| chr8_-_61407760 | 2.43 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr6_-_71609635 | 2.42 |
ENSMUST00000167220.4
ENSMUST00000205289.2 |
Kdm3a
|
lysine (K)-specific demethylase 3A |
| chr14_-_30909082 | 2.40 |
ENSMUST00000170253.8
|
Nisch
|
nischarin |
| chr14_+_25459630 | 2.39 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr18_+_44204457 | 2.38 |
ENSMUST00000068473.4
ENSMUST00000236634.2 |
Spink6
|
serine peptidase inhibitor, Kazal type 6 |
| chr14_-_86986541 | 2.32 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr18_-_70186503 | 2.27 |
ENSMUST00000117692.8
ENSMUST00000069749.9 |
Rab27b
|
RAB27B, member RAS oncogene family |
| chr7_+_127728712 | 2.26 |
ENSMUST00000033053.8
ENSMUST00000205460.2 |
Itgax
|
integrin alpha X |
| chr11_-_74615496 | 2.24 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr7_+_126376099 | 2.24 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr11_+_116324913 | 2.22 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr2_-_70655997 | 2.19 |
ENSMUST00000038584.9
|
Tlk1
|
tousled-like kinase 1 |
| chr9_+_117888124 | 2.16 |
ENSMUST00000123690.2
|
Azi2
|
5-azacytidine induced gene 2 |
| chr7_-_141925947 | 2.15 |
ENSMUST00000084412.6
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr11_-_69556888 | 2.14 |
ENSMUST00000108654.3
|
Cd68
|
CD68 antigen |
| chr15_-_58695379 | 2.14 |
ENSMUST00000072113.6
|
Tmem65
|
transmembrane protein 65 |
| chr4_-_133329479 | 2.12 |
ENSMUST00000057311.4
|
Sfn
|
stratifin |
| chr11_-_86648309 | 2.11 |
ENSMUST00000060766.16
ENSMUST00000103186.11 |
Cltc
|
clathrin, heavy polypeptide (Hc) |
| chr6_-_113411489 | 2.10 |
ENSMUST00000133348.2
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr15_-_38518406 | 2.09 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr11_-_69556904 | 2.09 |
ENSMUST00000018918.12
|
Cd68
|
CD68 antigen |
| chr7_-_30741512 | 2.06 |
ENSMUST00000159753.2
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr9_-_57168777 | 2.06 |
ENSMUST00000217657.2
|
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr2_-_102282844 | 2.04 |
ENSMUST00000099678.5
|
Fjx1
|
four jointed box 1 |
| chr10_-_79369584 | 2.00 |
ENSMUST00000218241.2
ENSMUST00000166804.2 ENSMUST00000063879.13 |
Plpp2
|
phospholipid phosphatase 2 |
| chr1_-_182345011 | 1.96 |
ENSMUST00000068505.10
|
Capn2
|
calpain 2 |
| chr3_-_92393193 | 1.92 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr12_-_84240781 | 1.90 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr12_+_83678987 | 1.90 |
ENSMUST00000048155.16
ENSMUST00000182618.8 ENSMUST00000183154.8 ENSMUST00000182036.8 ENSMUST00000182347.8 |
Rbm25
|
RNA binding motif protein 25 |
| chrX_-_7606445 | 1.89 |
ENSMUST00000128289.8
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr6_+_49013517 | 1.87 |
ENSMUST00000031840.10
|
Gpnmb
|
glycoprotein (transmembrane) nmb |
| chr14_-_32907446 | 1.81 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr7_+_120516967 | 1.81 |
ENSMUST00000207481.2
ENSMUST00000106483.4 ENSMUST00000033173.15 |
Polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr5_-_5315968 | 1.79 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr11_+_80274105 | 1.69 |
ENSMUST00000165565.8
ENSMUST00000188489.7 ENSMUST00000017567.14 ENSMUST00000108216.8 ENSMUST00000053740.15 |
Zfp207
|
zinc finger protein 207 |
| chr5_-_24597009 | 1.59 |
ENSMUST00000059401.7
|
Atg9b
|
autophagy related 9B |
| chr2_+_26481435 | 1.58 |
ENSMUST00000152988.9
ENSMUST00000149789.2 |
Egfl7
|
EGF-like domain 7 |
| chr17_+_75485791 | 1.56 |
ENSMUST00000135447.8
ENSMUST00000112516.8 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fos
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 43.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 5.3 | 16.0 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 3.8 | 11.3 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 3.6 | 10.7 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 3.5 | 20.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 3.1 | 21.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 2.8 | 87.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 2.7 | 18.7 | GO:0052205 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 2.6 | 13.0 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 2.3 | 13.8 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 2.2 | 10.8 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 2.0 | 9.8 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 1.9 | 15.6 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.9 | 9.5 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.9 | 5.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 1.9 | 7.5 | GO:0042097 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) |
| 1.7 | 5.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 1.7 | 8.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.7 | 30.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.6 | 6.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.5 | 7.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.5 | 23.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 1.4 | 8.4 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 1.4 | 2.8 | GO:0002649 | regulation of tolerance induction to self antigen(GO:0002649) |
| 1.3 | 3.9 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 1.3 | 3.9 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 1.2 | 3.7 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 1.1 | 11.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 1.0 | 27.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.9 | 21.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.8 | 2.5 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.8 | 2.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.8 | 5.6 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.8 | 6.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.7 | 12.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.7 | 13.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.7 | 5.7 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.7 | 2.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.7 | 2.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.7 | 9.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.7 | 6.7 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.7 | 5.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.7 | 9.9 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.6 | 2.6 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.6 | 15.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.6 | 3.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.5 | 9.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.5 | 3.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.5 | 2.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.5 | 4.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 | 1.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.4 | 1.8 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.4 | 5.7 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.4 | 2.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.4 | 4.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.4 | 5.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 2.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 2.9 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.4 | 11.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.3 | 2.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 8.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 3.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 2.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.3 | 13.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.3 | 1.2 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.3 | 4.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 0.8 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.3 | 3.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.3 | 1.3 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.3 | 4.9 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.2 | 2.1 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 1.6 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 2.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 2.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 2.4 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.2 | 1.6 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 1.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.2 | 0.5 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.2 | 9.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 1.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.2 | 1.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 3.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 5.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 2.7 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 7.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 2.5 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 1.1 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 0.5 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 4.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 8.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 7.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 18.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 1.4 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 3.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.6 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 2.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 6.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.0 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 9.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 3.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 5.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.2 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 12.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 2.8 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 2.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 1.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 3.5 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 1.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 5.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 1.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 2.7 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 4.8 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.2 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.8 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 3.3 | GO:0032720 | negative regulation of tumor necrosis factor production(GO:0032720) |
| 0.1 | 1.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 3.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 1.2 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 2.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 2.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.3 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 2.8 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 2.3 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 4.5 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 1.7 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 2.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0043305 | negative regulation of mast cell activation involved in immune response(GO:0033007) negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.6 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 2.8 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.0 | 1.9 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 1.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.9 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 1.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 1.5 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 1.5 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.6 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.0 | 0.2 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 1.4 | GO:0007030 | Golgi organization(GO:0007030) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.2 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 4.6 | 13.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 3.1 | 9.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 3.0 | 20.7 | GO:0005638 | lamin filament(GO:0005638) |
| 2.2 | 13.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 2.0 | 21.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.8 | 5.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 1.4 | 15.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.1 | 27.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.9 | 2.6 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.8 | 2.5 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.8 | 3.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.7 | 43.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.6 | 0.6 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.4 | 6.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.4 | 2.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.4 | 1.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.4 | 2.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 2.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.3 | 9.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 13.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 3.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 29.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 2.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 8.1 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 2.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 4.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 2.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 5.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 10.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 22.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 13.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.2 | 5.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 5.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.1 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 9.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 2.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.4 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 1.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 14.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 1.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 15.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 3.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 4.3 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 2.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 6.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 17.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 8.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 4.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 11.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 4.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 5.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 14.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.5 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 43.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 3.4 | 10.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 2.8 | 11.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 2.8 | 27.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.5 | 22.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 2.4 | 21.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 2.2 | 13.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 2.0 | 18.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.9 | 9.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.7 | 6.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.4 | 31.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 1.3 | 5.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 1.3 | 9.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.3 | 5.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 1.3 | 10.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.1 | 9.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.1 | 11.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.9 | 2.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.8 | 6.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.7 | 3.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.7 | 9.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.6 | 2.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.6 | 2.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.6 | 6.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.5 | 3.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 2.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.5 | 14.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.5 | 2.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.5 | 13.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.5 | 20.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.5 | 2.7 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 16.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 7.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 15.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.4 | 1.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 3.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 2.8 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.4 | 4.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 3.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 4.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 4.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 5.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 20.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 16.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 8.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 6.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 4.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 2.9 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.3 | 2.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 12.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 7.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 3.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 3.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 4.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 2.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 10.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 4.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 2.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 9.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 3.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.7 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 9.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 3.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 15.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 3.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 5.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 2.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 4.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 4.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 2.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 4.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 2.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.3 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 1.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 6.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 2.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 12.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.8 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 6.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 6.7 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 3.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.7 | 42.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.6 | 21.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.4 | 17.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.4 | 2.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 35.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.4 | 10.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.4 | 10.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 5.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 2.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 4.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 23.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 9.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 9.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 3.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 5.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 6.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 4.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 8.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 4.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 8.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 5.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 16.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 7.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.9 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 43.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.5 | 19.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.5 | 24.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 9.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.5 | 20.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 11.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 15.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 20.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.4 | 3.9 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 4.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 3.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 13.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 12.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 3.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 8.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.2 | 10.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 5.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 23.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.2 | 3.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 4.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 3.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.2 | 16.0 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.2 | 3.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 5.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 20.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 5.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 5.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 6.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 15.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 3.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 11.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 6.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.5 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 3.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 9.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |