Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
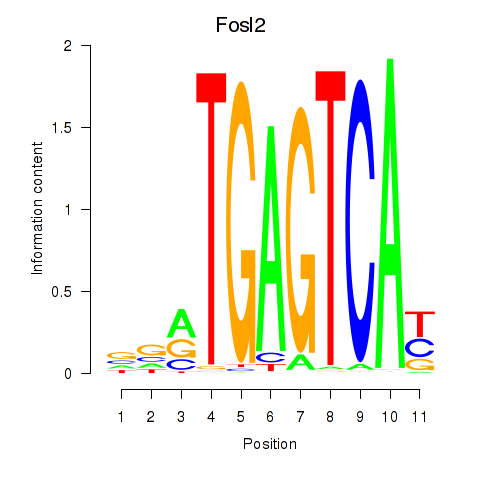
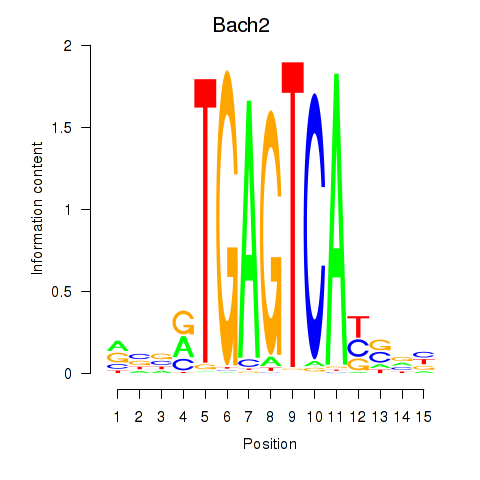
Results for Fosl2_Bach2
Z-value: 0.78
Transcription factors associated with Fosl2_Bach2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fosl2
|
ENSMUSG00000029135.11 | fos-like antigen 2 |
|
Bach2
|
ENSMUSG00000040270.17 | BTB and CNC homology, basic leucine zipper transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fosl2 | mm39_v1_chr5_+_32293145_32293201 | 0.28 | 1.0e-01 | Click! |
| Bach2 | mm39_v1_chr4_+_32238712_32238804 | 0.16 | 3.5e-01 | Click! |
Activity profile of Fosl2_Bach2 motif
Sorted Z-values of Fosl2_Bach2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102255999 | 19.49 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr1_-_132318039 | 15.43 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr8_+_95710977 | 10.23 |
ENSMUST00000093271.8
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr7_+_78563964 | 9.90 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr7_-_126398343 | 9.73 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_+_78563513 | 9.63 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr3_-_88410495 | 9.08 |
ENSMUST00000120377.8
ENSMUST00000029699.13 |
Lmna
|
lamin A |
| chr7_-_126398165 | 9.08 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr14_-_79539063 | 8.85 |
ENSMUST00000022595.8
|
Rgcc
|
regulator of cell cycle |
| chr2_+_164790139 | 8.42 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr1_+_174000304 | 8.13 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr7_+_78564062 | 7.84 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chrX_-_7537580 | 6.86 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr7_+_110371811 | 6.82 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr13_+_95833359 | 6.56 |
ENSMUST00000022182.5
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr3_+_93427791 | 6.36 |
ENSMUST00000029515.5
|
S100a11
|
S100 calcium binding protein A11 |
| chr8_+_95711037 | 5.77 |
ENSMUST00000211944.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_+_69361348 | 5.48 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr2_+_24226857 | 5.33 |
ENSMUST00000114487.9
ENSMUST00000142093.7 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr9_+_69360902 | 5.03 |
ENSMUST00000034756.15
ENSMUST00000123470.8 |
Anxa2
|
annexin A2 |
| chr1_+_75213258 | 4.87 |
ENSMUST00000185654.3
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr7_+_110372860 | 4.71 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr17_+_35268942 | 4.69 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr11_-_78950641 | 4.05 |
ENSMUST00000226282.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr8_+_94954280 | 3.84 |
ENSMUST00000109547.2
|
Nup93
|
nucleoporin 93 |
| chr7_+_126376353 | 3.69 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr7_+_110373447 | 3.57 |
ENSMUST00000147587.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr7_-_45570254 | 3.49 |
ENSMUST00000164119.3
|
Emp3
|
epithelial membrane protein 3 |
| chr7_-_45570538 | 3.47 |
ENSMUST00000210297.2
|
Emp3
|
epithelial membrane protein 3 |
| chr10_+_111309127 | 3.45 |
ENSMUST00000219143.2
|
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr9_-_44253588 | 3.32 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr6_-_115735935 | 3.29 |
ENSMUST00000072933.13
|
Tmem40
|
transmembrane protein 40 |
| chr10_+_75404482 | 3.27 |
ENSMUST00000134503.8
ENSMUST00000125770.8 ENSMUST00000128886.8 ENSMUST00000151212.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr2_+_84564394 | 3.21 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr16_+_35590745 | 3.21 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr11_+_61575245 | 3.19 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G |
| chrX_-_133483849 | 3.17 |
ENSMUST00000113213.2
ENSMUST00000033617.13 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr2_-_25114660 | 3.14 |
ENSMUST00000043584.5
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr11_-_109364424 | 3.11 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr7_-_19005721 | 3.09 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr7_-_45570674 | 3.03 |
ENSMUST00000210939.2
|
Emp3
|
epithelial membrane protein 3 |
| chr3_-_129763638 | 2.84 |
ENSMUST00000146340.2
ENSMUST00000153506.8 |
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chrX_-_165992311 | 2.81 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr6_-_56878854 | 2.77 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr9_+_51124983 | 2.73 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr3_+_95836558 | 2.68 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrX_-_165992145 | 2.59 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr1_+_75213171 | 2.59 |
ENSMUST00000187058.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr2_-_113883285 | 2.49 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr7_+_126376099 | 2.47 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr3_+_95836637 | 2.45 |
ENSMUST00000171368.8
ENSMUST00000168106.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_-_140501507 | 2.43 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr4_+_132857816 | 2.41 |
ENSMUST00000084241.12
ENSMUST00000138831.2 |
Wasf2
|
WASP family, member 2 |
| chr15_-_90563510 | 2.39 |
ENSMUST00000014777.9
ENSMUST00000064391.12 |
Cpne8
|
copine VIII |
| chr10_+_12966532 | 2.39 |
ENSMUST00000121646.8
ENSMUST00000121325.8 ENSMUST00000121766.8 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr6_+_42326760 | 2.38 |
ENSMUST00000203652.3
ENSMUST00000070635.13 |
Zyx
|
zyxin |
| chr9_-_44253630 | 2.31 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr4_-_6454262 | 2.30 |
ENSMUST00000029910.12
|
Nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr11_+_116422712 | 2.28 |
ENSMUST00000100201.10
|
Sphk1
|
sphingosine kinase 1 |
| chr11_+_116422570 | 2.26 |
ENSMUST00000106387.9
|
Sphk1
|
sphingosine kinase 1 |
| chr6_+_86605146 | 2.25 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr1_+_172328768 | 2.23 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr8_+_34222058 | 2.22 |
ENSMUST00000167264.8
ENSMUST00000187392.7 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr1_+_75213082 | 2.19 |
ENSMUST00000055223.14
ENSMUST00000082158.13 ENSMUST00000188346.7 |
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr8_+_34221861 | 2.13 |
ENSMUST00000170705.8
|
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr12_+_103354915 | 2.11 |
ENSMUST00000101094.9
ENSMUST00000021620.13 |
Otub2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr2_+_13578738 | 2.10 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr7_-_105131407 | 2.08 |
ENSMUST00000047040.4
|
Cavin3
|
caveolae associated 3 |
| chr10_+_75404464 | 2.04 |
ENSMUST00000145928.8
ENSMUST00000131565.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr15_-_38518406 | 2.02 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr11_-_95733235 | 2.02 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr17_+_46471950 | 2.01 |
ENSMUST00000024748.14
ENSMUST00000172170.8 |
Gtpbp2
|
GTP binding protein 2 |
| chr8_+_34222266 | 1.98 |
ENSMUST00000190675.2
ENSMUST00000171010.8 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr3_+_107803225 | 1.95 |
ENSMUST00000172247.8
ENSMUST00000167387.8 |
Gstm5
|
glutathione S-transferase, mu 5 |
| chr5_+_35156389 | 1.95 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr6_+_42326934 | 1.95 |
ENSMUST00000203401.3
ENSMUST00000164375.4 |
Zyx
|
zyxin |
| chrX_+_73468140 | 1.93 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr6_+_42326714 | 1.92 |
ENSMUST00000203846.3
|
Zyx
|
zyxin |
| chr1_+_75213114 | 1.89 |
ENSMUST00000188290.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr17_+_36152383 | 1.89 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr4_-_152533265 | 1.86 |
ENSMUST00000159840.8
ENSMUST00000105648.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr19_-_53020531 | 1.83 |
ENSMUST00000236008.2
ENSMUST00000237294.2 |
Xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr9_+_96078340 | 1.81 |
ENSMUST00000034982.16
ENSMUST00000188008.7 ENSMUST00000188750.7 ENSMUST00000185644.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr11_+_78079243 | 1.81 |
ENSMUST00000002128.14
ENSMUST00000150941.8 |
Rab34
|
RAB34, member RAS oncogene family |
| chr17_-_7050145 | 1.80 |
ENSMUST00000064234.7
|
Ezr
|
ezrin |
| chr17_-_48145466 | 1.78 |
ENSMUST00000066368.13
|
Mdfi
|
MyoD family inhibitor |
| chr10_+_128745214 | 1.78 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr3_+_107803137 | 1.77 |
ENSMUST00000004134.11
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr7_+_120516967 | 1.77 |
ENSMUST00000207481.2
ENSMUST00000106483.4 ENSMUST00000033173.15 |
Polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr4_-_126215462 | 1.73 |
ENSMUST00000102617.5
|
Adprs
|
ADP-ribosylserine hydrolase |
| chrX_-_103714653 | 1.72 |
ENSMUST00000042070.6
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr5_-_24597009 | 1.70 |
ENSMUST00000059401.7
|
Atg9b
|
autophagy related 9B |
| chr18_-_43610829 | 1.65 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr15_-_64184485 | 1.63 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr17_+_29170924 | 1.61 |
ENSMUST00000153462.8
|
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr3_+_92123106 | 1.59 |
ENSMUST00000074449.7
ENSMUST00000090871.3 |
Sprr2a1
Sprr2a2
|
small proline-rich protein 2A1 small proline-rich protein 2A2 |
| chrX_-_47543029 | 1.57 |
ENSMUST00000114958.8
|
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chrX_+_71006577 | 1.57 |
ENSMUST00000048790.7
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr18_+_82928959 | 1.55 |
ENSMUST00000171238.8
|
Zfp516
|
zinc finger protein 516 |
| chr11_-_78950698 | 1.54 |
ENSMUST00000141409.8
|
Ksr1
|
kinase suppressor of ras 1 |
| chr7_+_4463686 | 1.51 |
ENSMUST00000167298.2
ENSMUST00000171445.8 |
Eps8l1
|
EPS8-like 1 |
| chr14_+_79753055 | 1.51 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr7_+_27186335 | 1.48 |
ENSMUST00000008528.8
|
Sertad1
|
SERTA domain containing 1 |
| chr4_-_6454097 | 1.46 |
ENSMUST00000124344.2
|
Nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr17_+_29171655 | 1.42 |
ENSMUST00000117672.9
ENSMUST00000153831.9 |
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr10_+_87926932 | 1.41 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr8_-_61407760 | 1.39 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr6_-_143045493 | 1.32 |
ENSMUST00000204655.3
ENSMUST00000111758.9 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr17_-_57501170 | 1.29 |
ENSMUST00000005976.8
|
Tnfsf14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr18_-_38131766 | 1.25 |
ENSMUST00000236588.2
ENSMUST00000237272.2 ENSMUST00000236134.2 |
Arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr10_+_111309020 | 1.25 |
ENSMUST00000065917.16
ENSMUST00000219961.2 ENSMUST00000217908.2 |
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr5_+_35156454 | 1.25 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr5_-_21990170 | 1.24 |
ENSMUST00000115193.8
ENSMUST00000115192.2 ENSMUST00000115195.8 ENSMUST00000030771.12 |
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr4_+_43506966 | 1.22 |
ENSMUST00000030183.10
|
Car9
|
carbonic anhydrase 9 |
| chr10_+_127126643 | 1.21 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr1_+_75213044 | 1.20 |
ENSMUST00000188931.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr5_+_25452342 | 1.19 |
ENSMUST00000114950.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr7_-_16657825 | 1.18 |
ENSMUST00000019514.10
|
Calm3
|
calmodulin 3 |
| chr7_+_18817767 | 1.14 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr6_-_116650751 | 1.13 |
ENSMUST00000204576.2
ENSMUST00000203029.3 ENSMUST00000035842.7 |
Rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr14_-_56499690 | 1.11 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr16_+_33614715 | 1.10 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr10_-_128334515 | 1.10 |
ENSMUST00000026428.4
|
Myl6b
|
myosin, light polypeptide 6B |
| chr5_+_105563605 | 1.08 |
ENSMUST00000112707.3
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr2_+_30331839 | 1.08 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr15_+_84553801 | 1.06 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr2_-_101627999 | 1.05 |
ENSMUST00000171088.8
ENSMUST00000043845.14 |
Prr5l
|
proline rich 5 like |
| chr6_+_42326980 | 1.05 |
ENSMUST00000203849.2
|
Zyx
|
zyxin |
| chr19_+_4644425 | 1.05 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr11_+_78079631 | 1.05 |
ENSMUST00000056241.12
ENSMUST00000207728.2 |
Rab34
|
RAB34, member RAS oncogene family |
| chr3_-_10273628 | 1.04 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr6_+_135339543 | 1.04 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr1_-_75195127 | 1.02 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chrX_-_7606445 | 1.00 |
ENSMUST00000128289.8
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr6_+_42326528 | 0.99 |
ENSMUST00000203329.3
|
Zyx
|
zyxin |
| chr11_-_100098333 | 0.97 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chrX_-_100266032 | 0.96 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr1_-_52271455 | 0.95 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr19_-_46327024 | 0.95 |
ENSMUST00000236046.2
ENSMUST00000236980.2 ENSMUST00000235485.2 ENSMUST00000236061.2 ENSMUST00000236236.2 ENSMUST00000236768.2 ENSMUST00000236651.2 |
Cuedc2
|
CUE domain containing 2 |
| chr16_-_44379226 | 0.94 |
ENSMUST00000114634.3
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| chr4_+_109263820 | 0.92 |
ENSMUST00000124209.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chrX_+_95498965 | 0.92 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr18_-_70186503 | 0.92 |
ENSMUST00000117692.8
ENSMUST00000069749.9 |
Rab27b
|
RAB27B, member RAS oncogene family |
| chr6_+_56979285 | 0.92 |
ENSMUST00000079669.7
|
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr19_-_46327071 | 0.90 |
ENSMUST00000235977.2
ENSMUST00000167861.8 ENSMUST00000051234.9 ENSMUST00000236066.2 |
Cuedc2
|
CUE domain containing 2 |
| chrX_-_156275231 | 0.89 |
ENSMUST00000112529.8
|
Sms
|
spermine synthase |
| chr19_-_34143437 | 0.88 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr8_-_108315024 | 0.88 |
ENSMUST00000044106.6
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr12_-_84240781 | 0.87 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr2_+_151414524 | 0.86 |
ENSMUST00000028950.9
|
Sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr1_+_153625161 | 0.84 |
ENSMUST00000086209.10
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr7_+_126184108 | 0.84 |
ENSMUST00000039522.8
|
Apobr
|
apolipoprotein B receptor |
| chr11_-_69556888 | 0.83 |
ENSMUST00000108654.3
|
Cd68
|
CD68 antigen |
| chr19_-_8690330 | 0.83 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr1_+_153625243 | 0.82 |
ENSMUST00000182722.8
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr11_-_69556904 | 0.81 |
ENSMUST00000018918.12
|
Cd68
|
CD68 antigen |
| chr12_+_111725282 | 0.80 |
ENSMUST00000239017.2
ENSMUST00000084941.12 |
Klc1
|
kinesin light chain 1 |
| chr12_+_31315270 | 0.80 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chr10_-_76562002 | 0.79 |
ENSMUST00000001147.5
|
Col6a1
|
collagen, type VI, alpha 1 |
| chr14_-_86986541 | 0.77 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr10_+_115979787 | 0.77 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr8_-_25581354 | 0.76 |
ENSMUST00000125466.2
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr16_+_20470402 | 0.76 |
ENSMUST00000007212.9
ENSMUST00000232629.2 |
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr8_+_121215155 | 0.76 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr12_+_111725357 | 0.76 |
ENSMUST00000118471.8
ENSMUST00000122300.8 |
Klc1
|
kinesin light chain 1 |
| chr10_+_128247598 | 0.76 |
ENSMUST00000096386.13
|
Rnf41
|
ring finger protein 41 |
| chrX_+_161684563 | 0.75 |
ENSMUST00000112303.8
ENSMUST00000033727.14 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chrX_+_158623460 | 0.74 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr9_-_78388080 | 0.74 |
ENSMUST00000156988.2
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chrX_+_161684221 | 0.74 |
ENSMUST00000101095.9
|
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr16_+_36755338 | 0.73 |
ENSMUST00000023531.15
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr7_-_44861541 | 0.73 |
ENSMUST00000033057.9
|
Dkkl1
|
dickkopf-like 1 |
| chr3_+_64884839 | 0.72 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chrX_-_10083157 | 0.70 |
ENSMUST00000072393.9
ENSMUST00000044598.13 ENSMUST00000073392.11 ENSMUST00000115533.8 ENSMUST00000115532.2 |
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr13_-_61084358 | 0.70 |
ENSMUST00000225859.2
ENSMUST00000225167.2 ENSMUST00000021880.10 |
Gm49391
Ctla2a
|
predicted gene, 49391 cytotoxic T lymphocyte-associated protein 2 alpha |
| chr7_-_112946481 | 0.69 |
ENSMUST00000117577.8
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr19_-_32188413 | 0.68 |
ENSMUST00000151289.9
|
Sgms1
|
sphingomyelin synthase 1 |
| chr7_-_44861235 | 0.68 |
ENSMUST00000210741.2
ENSMUST00000209466.2 |
Dkkl1
|
dickkopf-like 1 |
| chr17_-_45903410 | 0.68 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr10_+_80765900 | 0.67 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr17_-_32569480 | 0.67 |
ENSMUST00000235265.2
ENSMUST00000236503.2 ENSMUST00000050214.9 |
Akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr2_+_156317416 | 0.64 |
ENSMUST00000029155.16
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr7_-_141925947 | 0.64 |
ENSMUST00000084412.6
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr10_+_128247492 | 0.64 |
ENSMUST00000171342.3
|
Rnf41
|
ring finger protein 41 |
| chr6_-_129484070 | 0.63 |
ENSMUST00000183258.8
ENSMUST00000182784.4 ENSMUST00000032265.13 ENSMUST00000162815.2 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr17_-_37178079 | 0.63 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr13_+_20295672 | 0.63 |
ENSMUST00000180626.2
|
Elmo1
|
engulfment and cell motility 1 |
| chr11_+_75546671 | 0.61 |
ENSMUST00000108431.3
|
Myo1c
|
myosin IC |
| chr1_-_75193937 | 0.61 |
ENSMUST00000186971.2
ENSMUST00000188593.2 |
Tuba4a
|
tubulin, alpha 4A |
| chr14_+_76714350 | 0.60 |
ENSMUST00000140251.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr17_-_45883421 | 0.59 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr12_+_31315227 | 0.59 |
ENSMUST00000169088.8
|
Lamb1
|
laminin B1 |
| chr13_-_91053440 | 0.59 |
ENSMUST00000109541.4
|
Atp6ap1l
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr11_+_80274105 | 0.58 |
ENSMUST00000165565.8
ENSMUST00000188489.7 ENSMUST00000017567.14 ENSMUST00000108216.8 ENSMUST00000053740.15 |
Zfp207
|
zinc finger protein 207 |
| chr17_-_15261701 | 0.58 |
ENSMUST00000097398.12
ENSMUST00000040746.8 ENSMUST00000097400.5 |
Dynlt2a1
Dynlt2a2
|
dynein light chain Tctex-type 2A1 dynein light chain Tctex-type 2A2 |
| chr4_+_128999325 | 0.58 |
ENSMUST00000106064.10
ENSMUST00000030575.15 ENSMUST00000030577.11 |
Tmem54
|
transmembrane protein 54 |
| chr18_-_38471962 | 0.57 |
ENSMUST00000139885.2
ENSMUST00000235590.2 ENSMUST00000237487.2 ENSMUST00000063814.15 |
Gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr12_+_111505216 | 0.57 |
ENSMUST00000050993.11
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr14_+_51181956 | 0.56 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr15_-_38518458 | 0.56 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fosl2_Bach2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 27.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 3.0 | 8.9 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 2.8 | 8.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.9 | 5.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 1.8 | 12.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.5 | 9.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.5 | 15.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.5 | 10.5 | GO:0052205 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 1.2 | 5.9 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.1 | 19.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.1 | 3.2 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.7 | 2.1 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 0.7 | 5.3 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.6 | 4.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.6 | 2.6 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.6 | 16.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.6 | 3.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.6 | 3.2 | GO:1905161 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.6 | 3.7 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.6 | 15.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 3.8 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.5 | 5.4 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.5 | 2.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.5 | 1.4 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.5 | 1.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.4 | 1.8 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.4 | 1.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.4 | 2.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.4 | 11.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 1.2 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.4 | 1.1 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.4 | 1.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.3 | 1.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 1.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 5.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.3 | 1.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.3 | 1.2 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.3 | 1.8 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.3 | 0.9 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 1.9 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 2.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.3 | 2.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 2.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 2.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 1.6 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 1.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 1.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 1.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 1.0 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 6.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.2 | 8.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 5.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 0.6 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 | 1.8 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.2 | 1.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.2 | 0.8 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 0.5 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.2 | 1.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 0.5 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.1 | 1.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.7 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.4 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 2.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 2.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.5 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 0.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.9 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.6 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 9.0 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 2.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.4 | GO:0010286 | heat acclimation(GO:0010286) regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 1.5 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.4 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.4 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 4.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.7 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 2.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 1.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.4 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 4.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.4 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 1.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.9 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 5.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.2 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 6.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.1 | 1.6 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 1.0 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.5 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 1.1 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.0 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.6 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.5 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.5 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0033007 | negative regulation of mast cell activation involved in immune response(GO:0033007) negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 2.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.4 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 1.7 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 1.2 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.5 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 7.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.7 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0018214 | protein carboxylation(GO:0018214) |
| 0.0 | 0.7 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 2.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.1 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0061723 | glycophagy(GO:0061723) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 2.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 2.6 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.6 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.6 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 1.4 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.5 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 2.1 | 6.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 1.5 | 16.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.4 | 8.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.3 | 9.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.9 | 22.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.8 | 2.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.7 | 2.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.6 | 1.8 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.6 | 1.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.5 | 27.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.4 | 1.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.4 | 2.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 1.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 3.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 1.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 2.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 0.9 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 16.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 10.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 1.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 4.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 1.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 3.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 4.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 5.1 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 2.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 3.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 1.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 7.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 27.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.9 | 18.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.9 | 5.6 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 1.8 | 5.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.6 | 6.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.5 | 15.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.2 | 10.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.9 | 3.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.8 | 18.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.6 | 2.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.5 | 2.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.5 | 12.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.5 | 2.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.5 | 4.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 5.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 1.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 1.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 5.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 2.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 1.2 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.3 | 1.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.3 | 1.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 1.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 6.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.2 | 9.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 8.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 7.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 0.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 1.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 15.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 1.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.7 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.1 | 1.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 3.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 5.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 2.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.7 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 3.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 5.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 1.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 1.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 5.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 1.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 3.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 7.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 2.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 1.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.9 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 2.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 3.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 3.8 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.0 | 0.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 4.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 9.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 11.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 8.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 7.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 17.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 12.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 12.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 15.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 29.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.4 | 18.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 4.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 5.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 9.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 9.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 7.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 6.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 3.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 3.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 15.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 4.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 2.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 2.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 2.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |