Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
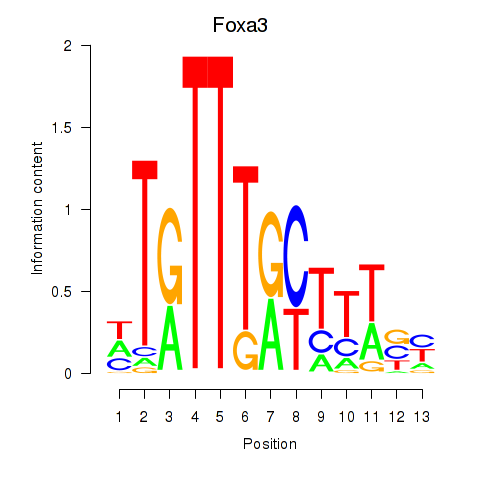
Results for Foxa3
Z-value: 1.06
Transcription factors associated with Foxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxa3
|
ENSMUSG00000040891.7 | forkhead box A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxa3 | mm39_v1_chr7_-_18757461_18757477 | 0.26 | 1.3e-01 | Click! |
Activity profile of Foxa3 motif
Sorted Z-values of Foxa3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_12894716 | 16.34 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr10_+_127734384 | 11.32 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr3_+_138121245 | 10.54 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr19_+_20579322 | 9.82 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr12_-_84497718 | 8.93 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr17_-_31363245 | 8.81 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr19_+_30210320 | 8.46 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr15_+_4756684 | 7.76 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr15_+_4756657 | 7.56 |
ENSMUST00000162585.8
|
C6
|
complement component 6 |
| chr19_-_20704896 | 7.22 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr3_-_20329823 | 7.11 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr15_-_96929086 | 7.02 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr9_+_46139878 | 6.73 |
ENSMUST00000034588.9
ENSMUST00000132155.2 |
Apoa1
|
apolipoprotein A-I |
| chr7_+_130633776 | 6.60 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr2_+_58645189 | 6.54 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr17_+_25097199 | 6.14 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr3_-_113166153 | 6.06 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr3_+_137983250 | 5.95 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr9_-_71070506 | 5.93 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr2_-_134396268 | 5.89 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr12_-_81014849 | 5.87 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr12_-_81014755 | 5.70 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr12_-_103739847 | 5.49 |
ENSMUST00000078869.6
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr2_+_58644922 | 5.28 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr1_+_88093726 | 5.20 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr4_-_6275629 | 5.13 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr15_+_3300249 | 4.89 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr4_+_104623505 | 4.76 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr12_+_8027767 | 4.69 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr12_+_8062331 | 4.63 |
ENSMUST00000171239.2
|
Apob
|
apolipoprotein B |
| chr12_+_8027640 | 4.52 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr2_+_67948057 | 4.39 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr14_-_30665232 | 4.21 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr18_-_39051695 | 4.16 |
ENSMUST00000040647.11
|
Fgf1
|
fibroblast growth factor 1 |
| chr7_-_105249308 | 4.10 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr6_-_23132977 | 3.86 |
ENSMUST00000031707.14
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr17_-_79292856 | 3.81 |
ENSMUST00000118991.2
|
Prkd3
|
protein kinase D3 |
| chr4_-_57916283 | 3.60 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr19_-_58442866 | 3.55 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr17_+_12597490 | 3.53 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr18_-_32271224 | 3.49 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
| chr11_-_69696428 | 3.49 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr15_-_54783357 | 3.44 |
ENSMUST00000167541.3
ENSMUST00000171545.9 ENSMUST00000041591.16 ENSMUST00000173516.8 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr3_+_20011405 | 3.44 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chrX_+_138464065 | 3.34 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr6_+_34575435 | 3.33 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr8_+_46944000 | 3.33 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr1_+_130793406 | 3.22 |
ENSMUST00000038829.7
|
Fcmr
|
Fc fragment of IgM receptor |
| chr10_-_95678786 | 3.21 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr3_-_146302343 | 3.17 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr4_-_63072367 | 3.16 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr11_+_78389913 | 3.12 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr3_+_20011251 | 3.07 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr5_+_90708962 | 3.06 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr9_-_103099262 | 2.99 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr8_+_46984016 | 2.96 |
ENSMUST00000152423.2
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr4_+_150938376 | 2.89 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr5_-_87054796 | 2.83 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr3_+_20011201 | 2.83 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr19_-_58444336 | 2.78 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr18_+_36797113 | 2.74 |
ENSMUST00000036765.8
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr10_-_53952686 | 2.63 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr2_-_104573179 | 2.61 |
ENSMUST00000028595.8
|
Depdc7
|
DEP domain containing 7 |
| chr3_+_107137924 | 2.60 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr11_+_78356523 | 2.29 |
ENSMUST00000001126.4
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr19_+_44980565 | 2.28 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr9_-_44714263 | 2.23 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr4_+_104770653 | 2.15 |
ENSMUST00000106803.9
ENSMUST00000106804.2 |
Fyb2
|
FYN binding protein 2 |
| chr3_-_148696155 | 2.04 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr4_+_148686985 | 2.03 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chrX_+_141011173 | 2.02 |
ENSMUST00000112914.8
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr8_-_26275182 | 1.95 |
ENSMUST00000038498.10
|
Bag4
|
BCL2-associated athanogene 4 |
| chr2_-_91025492 | 1.86 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chrX_+_141010919 | 1.79 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr15_-_5137975 | 1.67 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chr17_-_34847354 | 1.56 |
ENSMUST00000167097.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr17_+_29077385 | 1.56 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr1_+_172525613 | 1.54 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr4_+_101574601 | 1.53 |
ENSMUST00000102777.10
ENSMUST00000106921.9 ENSMUST00000037552.10 ENSMUST00000145024.2 |
Lepr
|
leptin receptor |
| chr11_+_108811168 | 1.49 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr7_+_18962252 | 1.49 |
ENSMUST00000063976.9
|
Opa3
|
optic atrophy 3 |
| chr3_+_85878376 | 1.46 |
ENSMUST00000238443.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr16_+_17149235 | 1.41 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr6_+_34686373 | 1.40 |
ENSMUST00000115021.8
|
Cald1
|
caldesmon 1 |
| chr15_-_5137951 | 1.38 |
ENSMUST00000141020.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr18_+_56533389 | 1.35 |
ENSMUST00000237355.2
ENSMUST00000237422.2 |
Gramd3
|
GRAM domain containing 3 |
| chr1_+_75119472 | 1.34 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chrX_-_55643429 | 1.31 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr12_-_108241597 | 1.27 |
ENSMUST00000222310.2
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chrX_+_108138965 | 1.25 |
ENSMUST00000033598.9
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr12_+_84498196 | 1.25 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr9_+_53212871 | 1.24 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr10_-_57408512 | 1.19 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr17_-_80203457 | 1.18 |
ENSMUST00000068282.7
ENSMUST00000112437.8 |
Atl2
|
atlastin GTPase 2 |
| chr12_+_59178072 | 1.17 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr1_+_75119419 | 1.14 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr18_-_84104507 | 1.12 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr2_+_140237229 | 1.11 |
ENSMUST00000110067.8
ENSMUST00000110063.8 ENSMUST00000110064.8 ENSMUST00000110062.8 ENSMUST00000043836.8 ENSMUST00000078027.12 |
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chr5_+_88731386 | 1.10 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr2_-_90309509 | 1.09 |
ENSMUST00000111495.9
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr2_+_164675697 | 1.08 |
ENSMUST00000143780.9
|
Ctsa
|
cathepsin A |
| chr11_+_108811626 | 1.05 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr11_-_69553390 | 1.05 |
ENSMUST00000129224.8
ENSMUST00000155200.8 |
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr15_-_99717956 | 1.04 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr7_+_18962301 | 1.04 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chrX_+_149377416 | 1.01 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr19_-_12773472 | 1.01 |
ENSMUST00000038627.9
|
Zfp91
|
zinc finger protein 91 |
| chr5_+_102629240 | 1.00 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr15_-_6904450 | 0.99 |
ENSMUST00000022746.13
ENSMUST00000176826.2 |
Osmr
|
oncostatin M receptor |
| chr4_+_45965327 | 0.99 |
ENSMUST00000107777.9
|
Tdrd7
|
tudor domain containing 7 |
| chr12_+_59178258 | 0.98 |
ENSMUST00000177162.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr10_-_57408585 | 0.97 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1 |
| chr2_+_153003212 | 0.95 |
ENSMUST00000089027.3
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr11_-_69553451 | 0.92 |
ENSMUST00000018905.12
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr5_+_88731366 | 0.91 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr17_+_43671314 | 0.89 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr14_-_40907106 | 0.89 |
ENSMUST00000077136.5
|
Sftpd
|
surfactant associated protein D |
| chr16_+_51851588 | 0.85 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr9_+_7692087 | 0.82 |
ENSMUST00000018767.8
|
Mmp7
|
matrix metallopeptidase 7 |
| chr12_-_32000209 | 0.82 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr6_+_29348068 | 0.81 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr11_-_54751738 | 0.81 |
ENSMUST00000144164.9
|
Lyrm7
|
LYR motif containing 7 |
| chr18_-_84104574 | 0.80 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr10_+_21868114 | 0.78 |
ENSMUST00000150089.8
ENSMUST00000100036.10 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_+_157353696 | 0.78 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr15_+_9071331 | 0.76 |
ENSMUST00000190591.10
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr14_-_121976273 | 0.76 |
ENSMUST00000212376.3
ENSMUST00000239077.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr12_-_32000169 | 0.72 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr2_-_77349909 | 0.72 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr15_-_3333003 | 0.69 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr14_+_103284413 | 0.67 |
ENSMUST00000022722.7
|
Acod1
|
aconitate decarboxylase 1 |
| chr13_+_89687915 | 0.67 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr9_+_32135540 | 0.67 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr7_+_92469100 | 0.64 |
ENSMUST00000107180.8
ENSMUST00000107179.2 |
Rab30
|
RAB30, member RAS oncogene family |
| chr13_+_24118417 | 0.64 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr17_+_34457868 | 0.61 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr12_-_87247082 | 0.59 |
ENSMUST00000037418.7
|
Tmed8
|
transmembrane p24 trafficking protein 8 |
| chr13_+_36301331 | 0.59 |
ENSMUST00000021857.13
|
Fars2
|
phenylalanine-tRNA synthetase 2 (mitochondrial) |
| chr12_+_112645237 | 0.58 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr1_-_168259264 | 0.58 |
ENSMUST00000176790.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr5_+_102629365 | 0.57 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr2_-_51039112 | 0.56 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr13_-_96678987 | 0.56 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chr3_-_146487102 | 0.56 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr11_-_46280336 | 0.53 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr11_+_96024612 | 0.51 |
ENSMUST00000167258.8
|
Ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr7_-_144761806 | 0.49 |
ENSMUST00000208788.2
|
Smim38
|
small integral membrane protein 38 |
| chr9_+_78020554 | 0.46 |
ENSMUST00000009972.6
ENSMUST00000117330.8 ENSMUST00000044551.8 |
Cilk1
|
ciliogenesis associated kinase 1 |
| chr2_-_70885877 | 0.44 |
ENSMUST00000090849.6
ENSMUST00000100037.9 ENSMUST00000112186.9 |
Mettl8
|
methyltransferase like 8 |
| chr17_-_43003135 | 0.44 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr4_+_137720326 | 0.43 |
ENSMUST00000139759.8
ENSMUST00000058133.10 ENSMUST00000105830.9 ENSMUST00000084215.12 |
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr17_-_31383976 | 0.43 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chr11_-_46280298 | 0.40 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr3_+_30800474 | 0.39 |
ENSMUST00000108262.10
|
Samd7
|
sterile alpha motif domain containing 7 |
| chr11_+_23615612 | 0.39 |
ENSMUST00000109525.8
ENSMUST00000020520.11 |
Pus10
|
pseudouridylate synthase 10 |
| chr15_+_54274151 | 0.38 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr11_-_46280281 | 0.37 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr7_+_101879176 | 0.37 |
ENSMUST00000120119.9
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr12_+_87247297 | 0.36 |
ENSMUST00000182869.2
|
Samd15
|
sterile alpha motif domain containing 15 |
| chr13_-_96678844 | 0.35 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr9_+_106247943 | 0.33 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr15_+_9071655 | 0.33 |
ENSMUST00000227682.3
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr1_+_32211792 | 0.33 |
ENSMUST00000027226.12
ENSMUST00000189878.2 ENSMUST00000188257.7 ENSMUST00000185666.2 |
Khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr5_+_38233901 | 0.32 |
ENSMUST00000146864.6
|
Stx18
|
syntaxin 18 |
| chr10_-_128579879 | 0.32 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr4_-_127247864 | 0.31 |
ENSMUST00000106090.8
ENSMUST00000060419.2 |
Gjb4
|
gap junction protein, beta 4 |
| chr11_+_96177449 | 0.31 |
ENSMUST00000049352.8
|
Hoxb7
|
homeobox B7 |
| chr10_-_128755127 | 0.30 |
ENSMUST00000149961.2
ENSMUST00000026406.14 |
Rdh5
|
retinol dehydrogenase 5 |
| chr2_+_65499097 | 0.29 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr10_+_25308466 | 0.28 |
ENSMUST00000219224.2
ENSMUST00000219166.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr6_-_99005212 | 0.28 |
ENSMUST00000177437.8
ENSMUST00000177229.8 ENSMUST00000124058.8 |
Foxp1
|
forkhead box P1 |
| chr3_-_27764522 | 0.28 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr18_+_40390013 | 0.28 |
ENSMUST00000096572.2
ENSMUST00000236889.2 |
Kctd16
|
potassium channel tetramerisation domain containing 16 |
| chr3_+_66893280 | 0.25 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr9_+_32135781 | 0.23 |
ENSMUST00000183121.2
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr14_+_32321341 | 0.23 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr8_-_48166955 | 0.22 |
ENSMUST00000212175.2
ENSMUST00000038738.7 |
Cdkn2aip
|
CDKN2A interacting protein |
| chr4_+_126156118 | 0.22 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chrX_-_107877909 | 0.22 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr2_+_130975417 | 0.20 |
ENSMUST00000110225.2
|
Gm11037
|
predicted gene 11037 |
| chr11_+_94455865 | 0.19 |
ENSMUST00000040418.9
|
Chad
|
chondroadherin |
| chr16_-_43484494 | 0.18 |
ENSMUST00000096065.6
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr3_-_92346078 | 0.18 |
ENSMUST00000062160.4
|
Sprr1b
|
small proline-rich protein 1B |
| chr3_+_66893031 | 0.17 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr12_-_32000534 | 0.17 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr3_+_33853941 | 0.17 |
ENSMUST00000099153.10
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr6_-_40590244 | 0.17 |
ENSMUST00000076565.3
|
Tas2r138
|
taste receptor, type 2, member 138 |
| chr9_+_107457316 | 0.16 |
ENSMUST00000093785.6
|
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr11_-_70560110 | 0.16 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr9_+_53757448 | 0.16 |
ENSMUST00000048485.7
|
Sln
|
sarcolipin |
| chr15_-_101778130 | 0.15 |
ENSMUST00000087996.7
|
Krt77
|
keratin 77 |
| chr9_-_101076198 | 0.15 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr10_+_19497740 | 0.14 |
ENSMUST00000036564.8
|
Il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr18_+_38809771 | 0.14 |
ENSMUST00000134388.2
ENSMUST00000148850.8 |
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene Rho GTPase activating protein 26 |
| chr8_-_106723026 | 0.14 |
ENSMUST00000227363.2
ENSMUST00000081998.13 |
Dpep2
|
dipeptidase 2 |
| chr9_-_79920131 | 0.13 |
ENSMUST00000217264.2
|
Filip1
|
filamin A interacting protein 1 |
| chr3_+_96127174 | 0.12 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21 |
| chr6_+_8948608 | 0.12 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr2_-_34261121 | 0.11 |
ENSMUST00000127353.3
ENSMUST00000141653.3 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr13_+_83672965 | 0.11 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxa3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 3.8 | 15.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 2.7 | 16.3 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 2.2 | 8.9 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 1.7 | 6.7 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 1.5 | 5.9 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.3 | 3.9 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.3 | 8.8 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 1.2 | 5.9 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 1.1 | 9.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.0 | 5.1 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 1.0 | 13.8 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.9 | 6.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.8 | 10.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.7 | 3.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.7 | 2.0 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.6 | 4.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.6 | 4.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.5 | 3.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.5 | 4.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 7.2 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.5 | 2.9 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.5 | 1.9 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.4 | 5.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 3.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.4 | 2.5 | GO:2000054 | regulation of mismatch repair(GO:0032423) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.4 | 3.5 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 11.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 1.5 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.4 | 3.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.4 | 5.7 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.3 | 0.7 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.3 | 4.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 2.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.3 | 10.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.3 | 0.8 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.3 | 1.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 6.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 3.1 | GO:0097421 | smooth muscle cell-matrix adhesion(GO:0061302) liver regeneration(GO:0097421) |
| 0.2 | 3.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.2 | 0.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 1.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 1.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 1.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 1.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 3.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 1.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 3.2 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 0.9 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 1.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 3.5 | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901028) |
| 0.1 | 7.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 2.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 2.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 1.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 2.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 3.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.9 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.7 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 2.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 11.4 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.1 | 1.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.1 | 0.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 2.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 1.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 2.1 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.5 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 4.2 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 1.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 2.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 2.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 3.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 1.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 4.5 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.6 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 4.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0034359 | mature chylomicron(GO:0034359) |
| 2.2 | 20.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.3 | 6.7 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.1 | 11.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.7 | 3.5 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.6 | 6.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 6.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.5 | 4.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 3.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 1.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 1.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.3 | 3.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 8.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 9.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 32.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 6.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.0 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 5.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 6.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.6 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 5.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 15.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 4.8 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 5.4 | 16.3 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 3.4 | 17.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 2.2 | 6.7 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 2.0 | 11.8 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.5 | 5.9 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 1.4 | 11.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.3 | 6.3 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 1.2 | 5.9 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 1.1 | 4.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.1 | 6.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.1 | 13.8 | GO:0035473 | lipase binding(GO:0035473) |
| 1.1 | 8.5 | GO:0005534 | galactose binding(GO:0005534) |
| 0.9 | 6.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.8 | 9.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.8 | 3.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.7 | 3.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.7 | 3.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.6 | 3.2 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.6 | 8.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.6 | 11.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 3.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.3 | 1.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.3 | 4.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 3.6 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.2 | 6.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 2.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 3.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 8.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 9.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 4.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 2.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 10.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.2 | 1.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 5.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 3.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.3 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 2.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 5.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 2.9 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 10.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 5.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 4.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 17.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 3.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 6.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 5.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 8.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 12.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 9.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 8.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 20.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.5 | 12.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.2 | 16.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.1 | 11.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.9 | 20.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.7 | 20.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 11.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 5.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 9.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 5.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 8.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 3.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 5.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 6.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 9.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 4.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 6.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 4.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 8.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |