Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Foxd2
Z-value: 0.48
Transcription factors associated with Foxd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd2
|
ENSMUSG00000055210.5 | forkhead box D2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxd2 | mm39_v1_chr4_-_114766070_114766111 | 0.01 | 9.3e-01 | Click! |
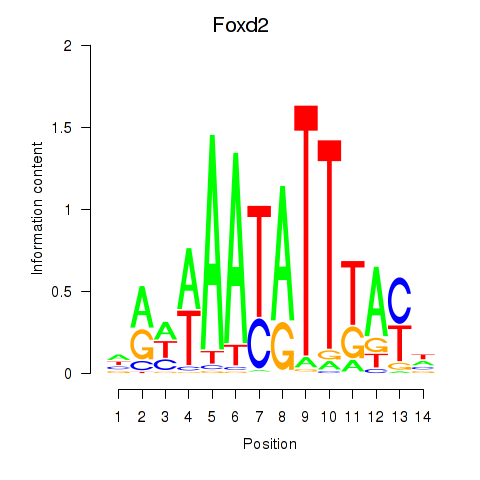
Activity profile of Foxd2 motif
Sorted Z-values of Foxd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_107456731 | 3.02 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr9_+_96140781 | 2.76 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr14_+_80237691 | 2.74 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr14_-_70867588 | 2.55 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr11_+_95227836 | 2.37 |
ENSMUST00000037502.7
|
Fam117a
|
family with sequence similarity 117, member A |
| chr6_+_38895902 | 1.74 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr7_+_130633776 | 1.58 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr15_-_98505508 | 1.52 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr13_-_97235745 | 1.47 |
ENSMUST00000071118.7
|
Gm6169
|
predicted gene 6169 |
| chr15_-_101833160 | 1.46 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr2_+_24235300 | 1.34 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr2_-_25498459 | 1.21 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr4_-_131695135 | 1.13 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr9_+_96140750 | 1.11 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr17_+_29309942 | 1.07 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr7_-_108769719 | 1.06 |
ENSMUST00000208136.2
ENSMUST00000036992.9 |
Lmo1
|
LIM domain only 1 |
| chr10_-_88440869 | 1.01 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr10_-_88440996 | 1.00 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr2_-_62313981 | 0.96 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr19_-_53577499 | 0.94 |
ENSMUST00000095978.5
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chr13_+_94954202 | 0.89 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr18_-_6516089 | 0.88 |
ENSMUST00000115870.9
|
Epc1
|
enhancer of polycomb homolog 1 |
| chr7_-_13856967 | 0.78 |
ENSMUST00000098809.4
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr13_-_20008397 | 0.74 |
ENSMUST00000222664.2
ENSMUST00000065335.3 |
Gpr141
|
G protein-coupled receptor 141 |
| chr4_+_108704982 | 0.73 |
ENSMUST00000102738.4
|
Kti12
|
KTI12 homolog, chromatin associated |
| chr6_+_142244145 | 0.73 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr11_-_75330302 | 0.72 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr11_-_107080150 | 0.70 |
ENSMUST00000106757.8
ENSMUST00000018577.8 |
Nol11
|
nucleolar protein 11 |
| chr18_+_37637317 | 0.69 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr11_-_75330415 | 0.68 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr6_-_119925387 | 0.68 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr17_+_8144822 | 0.65 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr10_-_126866682 | 0.64 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr2_+_87853118 | 0.63 |
ENSMUST00000214438.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr7_+_97480125 | 0.63 |
ENSMUST00000206351.2
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr3_+_75982890 | 0.61 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr19_-_8691460 | 0.61 |
ENSMUST00000206560.2
ENSMUST00000205538.2 |
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr9_-_90152759 | 0.60 |
ENSMUST00000041767.14
ENSMUST00000128874.3 |
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr5_+_149601688 | 0.59 |
ENSMUST00000100404.6
|
B3glct
|
beta-3-glucosyltransferase |
| chr6_-_16898440 | 0.58 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr1_+_171668173 | 0.58 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr18_+_44467133 | 0.56 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chr10_-_126866658 | 0.51 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr1_-_173594475 | 0.50 |
ENSMUST00000111214.4
|
Ifi204
|
interferon activated gene 204 |
| chrX_+_132751729 | 0.49 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr1_-_173018204 | 0.48 |
ENSMUST00000215878.2
ENSMUST00000201132.3 |
Olfr1406
|
olfactory receptor 1406 |
| chr3_-_72875187 | 0.47 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr12_+_38833454 | 0.47 |
ENSMUST00000161980.8
ENSMUST00000160701.8 |
Etv1
|
ets variant 1 |
| chrX_-_100312629 | 0.46 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr12_+_38833501 | 0.46 |
ENSMUST00000159334.8
|
Etv1
|
ets variant 1 |
| chr3_-_144738526 | 0.45 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr16_-_45313244 | 0.43 |
ENSMUST00000232138.2
|
Gm609
|
predicted gene 609 |
| chr14_-_50380137 | 0.43 |
ENSMUST00000213685.2
|
Olfr728
|
olfactory receptor 728 |
| chr2_-_87868043 | 0.40 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73 |
| chr16_-_45313324 | 0.39 |
ENSMUST00000114585.3
|
Gm609
|
predicted gene 609 |
| chr11_+_75541324 | 0.38 |
ENSMUST00000102505.10
|
Myo1c
|
myosin IC |
| chr10_+_57521930 | 0.38 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr1_-_173105344 | 0.38 |
ENSMUST00000111224.5
|
Mptx2
|
mucosal pentraxin 2 |
| chrX_+_162922317 | 0.37 |
ENSMUST00000112271.10
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr1_+_88334678 | 0.36 |
ENSMUST00000027518.12
|
Spp2
|
secreted phosphoprotein 2 |
| chr4_-_45108038 | 0.34 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr7_-_34012956 | 0.34 |
ENSMUST00000108074.8
|
Garre1
|
granule associated Rac and RHOG effector 1 |
| chr6_-_57827328 | 0.33 |
ENSMUST00000203310.3
ENSMUST00000203488.3 |
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr1_-_173740467 | 0.33 |
ENSMUST00000009340.10
|
Ifi211
|
interferon activated gene 211 |
| chr1_-_126758369 | 0.33 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr10_+_57521958 | 0.32 |
ENSMUST00000177473.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr2_-_11608003 | 0.31 |
ENSMUST00000040314.12
|
Rbm17
|
RNA binding motif protein 17 |
| chr2_+_132532040 | 0.30 |
ENSMUST00000148271.8
ENSMUST00000110132.3 |
Shld1
|
shieldin complex subunit 1 |
| chr7_-_132616977 | 0.29 |
ENSMUST00000169570.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr3_+_89427458 | 0.29 |
ENSMUST00000000811.8
|
Kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr7_+_28834276 | 0.28 |
ENSMUST00000161522.8
ENSMUST00000204845.3 ENSMUST00000205027.3 ENSMUST00000204194.3 ENSMUST00000203070.3 ENSMUST00000203380.3 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr3_-_75177378 | 0.28 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr9_-_78016302 | 0.27 |
ENSMUST00000001402.14
|
Fbxo9
|
f-box protein 9 |
| chr7_-_34012934 | 0.26 |
ENSMUST00000206399.2
|
Garre1
|
granule associated Rac and RHOG effector 1 |
| chr13_-_98152768 | 0.25 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr10_-_18887701 | 0.25 |
ENSMUST00000105527.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chrX_-_104973003 | 0.24 |
ENSMUST00000130980.2
ENSMUST00000113573.8 |
Atrx
|
ATRX, chromatin remodeler |
| chr7_-_85065128 | 0.24 |
ENSMUST00000171213.3
ENSMUST00000233336.2 |
Vmn2r69
|
vomeronasal 2, receptor 69 |
| chr11_+_98689479 | 0.24 |
ENSMUST00000037930.13
|
Msl1
|
male specific lethal 1 |
| chr9_+_38340751 | 0.24 |
ENSMUST00000216502.3
ENSMUST00000216644.2 |
Olfr901
|
olfactory receptor 901 |
| chr2_-_87570322 | 0.24 |
ENSMUST00000214573.2
|
Olfr1138
|
olfactory receptor 1138 |
| chr16_-_19132814 | 0.23 |
ENSMUST00000216157.2
|
Olfr164
|
olfactory receptor 164 |
| chr2_-_110591909 | 0.23 |
ENSMUST00000140777.2
|
Ano3
|
anoctamin 3 |
| chr4_-_136626073 | 0.23 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr3_-_123483772 | 0.23 |
ENSMUST00000172537.3
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr2_-_86061745 | 0.22 |
ENSMUST00000216056.2
|
Olfr1047
|
olfactory receptor 1047 |
| chr7_-_13723513 | 0.22 |
ENSMUST00000165167.8
ENSMUST00000108520.4 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr2_-_87881473 | 0.22 |
ENSMUST00000183862.3
|
Olfr1162
|
olfactory receptor 1162 |
| chr10_+_42736771 | 0.21 |
ENSMUST00000105494.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr1_-_118239146 | 0.21 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr4_+_128621378 | 0.21 |
ENSMUST00000106079.10
ENSMUST00000133439.8 |
Phc2
|
polyhomeotic 2 |
| chr10_+_128067964 | 0.21 |
ENSMUST00000125289.8
ENSMUST00000105242.8 |
Timeless
|
timeless circadian clock 1 |
| chr12_-_41536430 | 0.20 |
ENSMUST00000043884.6
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr5_+_18167547 | 0.18 |
ENSMUST00000030561.9
|
Gnat3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr9_-_95632387 | 0.18 |
ENSMUST00000189137.7
ENSMUST00000053785.10 |
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr9_-_55190902 | 0.18 |
ENSMUST00000164721.8
|
Nrg4
|
neuregulin 4 |
| chr17_-_56583715 | 0.17 |
ENSMUST00000058136.9
|
Ticam1
|
toll-like receptor adaptor molecule 1 |
| chr16_-_4698148 | 0.17 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr15_+_100052260 | 0.17 |
ENSMUST00000023768.14
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr7_-_126795096 | 0.17 |
ENSMUST00000206026.2
ENSMUST00000205321.2 ENSMUST00000206587.2 ENSMUST00000205316.2 ENSMUST00000166791.8 |
Cd2bp2
|
CD2 cytoplasmic tail binding protein 2 |
| chr18_+_68470616 | 0.16 |
ENSMUST00000172148.4
ENSMUST00000239469.2 |
Mc5r
|
melanocortin 5 receptor |
| chr19_-_31742427 | 0.16 |
ENSMUST00000065067.14
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr5_+_107645626 | 0.15 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr3_+_7494108 | 0.15 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr7_-_126795040 | 0.15 |
ENSMUST00000035771.5
|
Cd2bp2
|
CD2 cytoplasmic tail binding protein 2 |
| chr18_+_73996743 | 0.15 |
ENSMUST00000134847.2
|
Mro
|
maestro |
| chr18_-_60724855 | 0.15 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr2_-_73143045 | 0.15 |
ENSMUST00000058615.10
|
Cir1
|
corepressor interacting with RBPJ, 1 |
| chr9_+_72600721 | 0.15 |
ENSMUST00000238315.2
|
Nedd4
|
neural precursor cell expressed, developmentally down-regulated 4 |
| chr2_-_86008164 | 0.14 |
ENSMUST00000215171.2
|
Olfr1044
|
olfactory receptor 1044 |
| chr18_+_37858753 | 0.14 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr2_-_87504008 | 0.14 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr12_+_65122355 | 0.14 |
ENSMUST00000058889.5
|
Fancm
|
Fanconi anemia, complementation group M |
| chr3_+_114874614 | 0.14 |
ENSMUST00000051309.9
|
Olfm3
|
olfactomedin 3 |
| chr6_+_11926757 | 0.13 |
ENSMUST00000133776.2
|
Phf14
|
PHD finger protein 14 |
| chrX_+_108240356 | 0.13 |
ENSMUST00000139259.2
ENSMUST00000060013.4 |
Gm6377
|
predicted gene 6377 |
| chr3_-_14676261 | 0.13 |
ENSMUST00000108365.4
|
Rbis
|
ribosomal biogenesis factor |
| chr11_+_29413734 | 0.13 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr4_-_82768958 | 0.13 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chrX_+_158480304 | 0.13 |
ENSMUST00000123433.8
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr2_-_85632888 | 0.12 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr14_+_75368532 | 0.12 |
ENSMUST00000143539.8
ENSMUST00000134114.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr14_+_50741057 | 0.12 |
ENSMUST00000217437.2
ENSMUST00000213935.2 |
Olfr742
|
olfactory receptor 742 |
| chr12_-_101924407 | 0.12 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chrX_+_67805443 | 0.12 |
ENSMUST00000069731.12
ENSMUST00000114647.8 |
Fmr1nb
|
Fmr1 neighbor |
| chr2_+_14393245 | 0.11 |
ENSMUST00000133258.2
|
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr14_+_50656083 | 0.11 |
ENSMUST00000216949.2
|
Olfr739
|
olfactory receptor 739 |
| chr1_-_126758520 | 0.11 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr5_+_20433169 | 0.10 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_-_58009191 | 0.10 |
ENSMUST00000159826.2
ENSMUST00000164963.8 |
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr14_+_54113415 | 0.10 |
ENSMUST00000180938.3
|
Trav21-dv12
|
T cell receptor alpha variable 21-DV12 |
| chr17_+_38110779 | 0.09 |
ENSMUST00000215168.2
ENSMUST00000216478.2 |
Olfr124
|
olfactory receptor 124 |
| chr2_-_172782089 | 0.09 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
| chr11_+_58549642 | 0.09 |
ENSMUST00000214392.2
|
Olfr322
|
olfactory receptor 322 |
| chr1_-_44140820 | 0.09 |
ENSMUST00000152239.8
|
Tex30
|
testis expressed 30 |
| chrY_-_70026134 | 0.09 |
ENSMUST00000188554.7
ENSMUST00000191151.2 |
Gm28079
|
predicted gene 28079 |
| chr14_+_50618620 | 0.09 |
ENSMUST00000215263.2
ENSMUST00000213402.2 ENSMUST00000213755.2 ENSMUST00000215227.2 |
Olfr736
|
olfactory receptor 736 |
| chr6_+_136495784 | 0.08 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr9_+_37995368 | 0.08 |
ENSMUST00000212502.4
|
Olfr887
|
olfactory receptor 887 |
| chr16_-_58583539 | 0.08 |
ENSMUST00000214139.2
|
Olfr172
|
olfactory receptor 172 |
| chr18_+_89224219 | 0.07 |
ENSMUST00000236835.2
|
Cd226
|
CD226 antigen |
| chr11_+_78811613 | 0.07 |
ENSMUST00000018610.7
|
Nos2
|
nitric oxide synthase 2, inducible |
| chr1_-_44141503 | 0.07 |
ENSMUST00000128190.8
ENSMUST00000147571.8 ENSMUST00000027215.12 ENSMUST00000147661.8 |
Tex30
|
testis expressed 30 |
| chr10_+_129320621 | 0.06 |
ENSMUST00000213236.2
ENSMUST00000213992.2 |
Olfr789
|
olfactory receptor 789 |
| chr6_-_57992144 | 0.06 |
ENSMUST00000228070.2
ENSMUST00000228040.2 |
Vmn1r26
|
vomeronasal 1 receptor 26 |
| chr10_+_129907079 | 0.06 |
ENSMUST00000216879.2
|
Olfr822
|
olfactory receptor 822 |
| chrX_-_49517375 | 0.06 |
ENSMUST00000215270.2
|
Olfr1324
|
olfactory receptor 1324 |
| chr5_-_87847268 | 0.06 |
ENSMUST00000196869.5
ENSMUST00000199624.5 ENSMUST00000198057.5 ENSMUST00000082370.10 |
Csn2
|
casein beta |
| chr2_-_45000389 | 0.05 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr8_-_45715049 | 0.05 |
ENSMUST00000034064.5
|
F11
|
coagulation factor XI |
| chr9_+_38725910 | 0.05 |
ENSMUST00000213164.2
|
Olfr922
|
olfactory receptor 922 |
| chr3_-_144511566 | 0.04 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr6_+_57086439 | 0.04 |
ENSMUST00000228270.2
|
Vmn1r10
|
vomeronasal 1 receptor 10 |
| chr2_-_111779785 | 0.04 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr5_-_110220379 | 0.04 |
ENSMUST00000210275.2
|
Gm17655
|
predicted gene, 17655 |
| chr6_-_3494587 | 0.04 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr16_-_58620631 | 0.04 |
ENSMUST00000206205.3
|
Olfr173
|
olfactory receptor 173 |
| chr2_+_14393127 | 0.04 |
ENSMUST00000114731.8
ENSMUST00000082290.8 |
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr14_-_75368321 | 0.03 |
ENSMUST00000022574.5
|
Lrrc63
|
leucine rich repeat containing 63 |
| chr5_-_23889607 | 0.03 |
ENSMUST00000197985.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chrX_+_82898974 | 0.03 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy |
| chrX_-_156198282 | 0.02 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr10_-_130050272 | 0.02 |
ENSMUST00000213568.2
|
Olfr827
|
olfactory receptor 827 |
| chrX_-_50031587 | 0.02 |
ENSMUST00000060650.7
|
Frmd7
|
FERM domain containing 7 |
| chr7_+_99384352 | 0.02 |
ENSMUST00000098264.2
|
Olfr520
|
olfactory receptor 520 |
| chr14_+_102077937 | 0.01 |
ENSMUST00000159026.8
|
Lmo7
|
LIM domain only 7 |
| chr6_-_129600812 | 0.01 |
ENSMUST00000168919.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr1_-_37535170 | 0.01 |
ENSMUST00000148047.2
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chrX_-_104972938 | 0.01 |
ENSMUST00000198448.5
ENSMUST00000199233.5 ENSMUST00000134507.8 ENSMUST00000154866.8 ENSMUST00000128968.8 ENSMUST00000134381.8 ENSMUST00000150914.8 |
Atrx
|
ATRX, chromatin remodeler |
| chr11_+_104983022 | 0.01 |
ENSMUST00000021029.6
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr12_-_111946560 | 0.01 |
ENSMUST00000190680.2
|
Rd3l
|
retinal degeneration 3-like |
| chr5_+_24598633 | 0.01 |
ENSMUST00000138168.3
ENSMUST00000115077.8 |
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr9_-_58026090 | 0.00 |
ENSMUST00000098681.4
ENSMUST00000098682.10 ENSMUST00000215944.2 |
Ccdc33
|
coiled-coil domain containing 33 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.4 | 2.6 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.3 | 1.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 1.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.6 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.2 | 0.9 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 1.4 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.2 | 0.6 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.2 | 1.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.7 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 2.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 1.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 1.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.4 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.1 | 0.7 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.3 | GO:0002632 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.2 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 1.1 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 0.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 1.1 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.7 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.2 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 2.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0060369 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 1.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 0.7 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.2 | 1.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 2.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 1.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 3.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.4 | 1.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 1.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.0 | GO:0050656 | alcohol sulfotransferase activity(GO:0004027) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 2.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 3.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 1.7 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |