Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
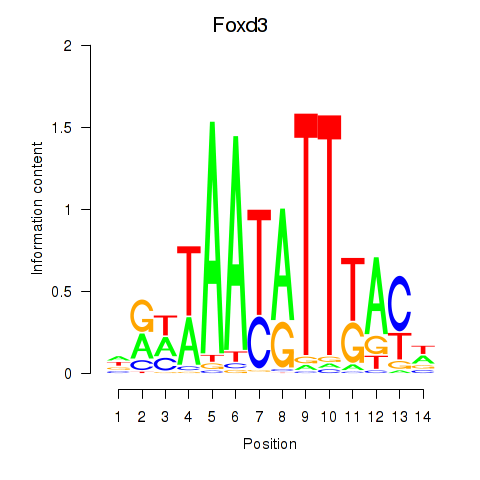
Results for Foxd3
Z-value: 0.56
Transcription factors associated with Foxd3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd3
|
ENSMUSG00000067261.5 | forkhead box D3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxd3 | mm39_v1_chr4_+_99544536_99544577 | 0.03 | 8.7e-01 | Click! |
Activity profile of Foxd3 motif
Sorted Z-values of Foxd3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_103739847 | 4.54 |
ENSMUST00000078869.6
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr10_+_93324624 | 4.07 |
ENSMUST00000129421.8
|
Hal
|
histidine ammonia lyase |
| chr12_-_103871146 | 3.62 |
ENSMUST00000074051.6
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr10_-_95678786 | 3.04 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr3_+_146302832 | 3.00 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr10_-_95678748 | 2.99 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr5_-_31453206 | 2.96 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chr1_+_88066086 | 2.92 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr13_-_56696310 | 2.85 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr13_-_56696222 | 2.68 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr9_-_71070506 | 2.46 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr12_-_103923145 | 2.30 |
ENSMUST00000085054.5
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr19_-_38113696 | 2.07 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr9_-_50466470 | 1.93 |
ENSMUST00000119103.2
|
Bco2
|
beta-carotene oxygenase 2 |
| chr6_-_21851827 | 1.83 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr7_+_67305162 | 1.68 |
ENSMUST00000107470.2
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr17_-_31855605 | 1.61 |
ENSMUST00000151718.3
ENSMUST00000135425.9 ENSMUST00000155814.8 |
Cbs
|
cystathionine beta-synthase |
| chr14_-_45626237 | 1.17 |
ENSMUST00000227865.2
ENSMUST00000226856.2 ENSMUST00000226276.2 ENSMUST00000046191.9 |
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr9_+_50466127 | 1.07 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr2_+_152511381 | 1.06 |
ENSMUST00000125366.8
ENSMUST00000109825.8 ENSMUST00000089059.9 ENSMUST00000079247.4 |
H13
|
histocompatibility 13 |
| chr3_-_146302343 | 0.99 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr9_-_20791012 | 0.87 |
ENSMUST00000043726.8
|
Angptl6
|
angiopoietin-like 6 |
| chr13_+_41040657 | 0.85 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr1_+_88334678 | 0.83 |
ENSMUST00000027518.12
|
Spp2
|
secreted phosphoprotein 2 |
| chr3_-_82957104 | 0.71 |
ENSMUST00000048246.5
|
Fgb
|
fibrinogen beta chain |
| chr7_-_99629637 | 0.70 |
ENSMUST00000080817.6
|
Rnf169
|
ring finger protein 169 |
| chr18_+_44467133 | 0.67 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chr17_+_29309942 | 0.63 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr12_-_103597663 | 0.63 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr12_-_110807330 | 0.60 |
ENSMUST00000177224.2
ENSMUST00000084974.11 ENSMUST00000070565.15 |
Mok
|
MOK protein kinase |
| chr16_-_4698148 | 0.59 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr2_-_18397547 | 0.56 |
ENSMUST00000091418.12
ENSMUST00000166495.8 |
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr17_-_35265702 | 0.55 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr8_-_45715049 | 0.51 |
ENSMUST00000034064.5
|
F11
|
coagulation factor XI |
| chr17_+_28910393 | 0.50 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr11_-_34048538 | 0.48 |
ENSMUST00000223852.2
|
4930469K13Rik
|
RIKEN cDNA 4930469K13 gene |
| chr18_+_37646674 | 0.48 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
| chr15_-_6904450 | 0.45 |
ENSMUST00000022746.13
ENSMUST00000176826.2 |
Osmr
|
oncostatin M receptor |
| chr6_-_131224305 | 0.42 |
ENSMUST00000032306.15
ENSMUST00000088867.7 |
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr17_+_28910302 | 0.37 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr15_+_92495007 | 0.35 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr6_+_142244145 | 0.31 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr5_+_148202011 | 0.31 |
ENSMUST00000110515.9
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr2_-_17465410 | 0.30 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr11_-_75330302 | 0.28 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr5_+_148202117 | 0.27 |
ENSMUST00000110514.8
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr10_-_89457115 | 0.24 |
ENSMUST00000020102.14
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr17_+_93506435 | 0.24 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr10_-_88440869 | 0.24 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr18_+_37858753 | 0.23 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr8_-_88686188 | 0.22 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr12_+_111937978 | 0.22 |
ENSMUST00000079009.11
|
Tdrd9
|
tudor domain containing 9 |
| chr17_+_93506590 | 0.22 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr13_+_23715220 | 0.21 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr6_+_77219698 | 0.19 |
ENSMUST00000161677.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chrX_+_163156359 | 0.18 |
ENSMUST00000033751.8
|
Vegfd
|
vascular endothelial growth factor D |
| chr1_+_6805048 | 0.18 |
ENSMUST00000139838.8
|
St18
|
suppression of tumorigenicity 18 |
| chr11_-_75330415 | 0.17 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr2_+_124978518 | 0.17 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr5_+_148202075 | 0.16 |
ENSMUST00000071878.12
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr2_+_181408833 | 0.15 |
ENSMUST00000108756.8
|
Myt1
|
myelin transcription factor 1 |
| chrX_-_156198282 | 0.14 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr6_+_77219627 | 0.14 |
ENSMUST00000159616.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr2_+_131792774 | 0.13 |
ENSMUST00000110170.8
ENSMUST00000110172.9 ENSMUST00000110171.9 |
Prnd
|
prion like protein doppel |
| chr14_-_45626198 | 0.13 |
ENSMUST00000226590.2
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr6_-_141957140 | 0.13 |
ENSMUST00000181791.8
ENSMUST00000181628.8 |
Gm6614
|
predicted gene 6614 |
| chr2_+_51928017 | 0.12 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr12_+_103564479 | 0.12 |
ENSMUST00000190151.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chrX_+_132751729 | 0.11 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr2_+_14393127 | 0.11 |
ENSMUST00000114731.8
ENSMUST00000082290.8 |
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr2_+_131792931 | 0.11 |
ENSMUST00000110169.2
|
Prnd
|
prion like protein doppel |
| chr15_-_99185050 | 0.10 |
ENSMUST00000109100.2
|
Fam186b
|
family with sequence similarity 186, member B |
| chr11_-_99884818 | 0.10 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr7_+_91321694 | 0.09 |
ENSMUST00000238608.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr10_-_88440996 | 0.09 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr9_+_37995368 | 0.09 |
ENSMUST00000212502.4
|
Olfr887
|
olfactory receptor 887 |
| chr10_+_66932235 | 0.09 |
ENSMUST00000174317.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr2_+_181409075 | 0.08 |
ENSMUST00000108757.9
|
Myt1
|
myelin transcription factor 1 |
| chr10_-_128579879 | 0.08 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr1_+_53100796 | 0.07 |
ENSMUST00000027269.7
ENSMUST00000191197.2 |
Mstn
|
myostatin |
| chr2_+_124978612 | 0.07 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr5_-_86475595 | 0.07 |
ENSMUST00000122377.2
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr15_-_101833160 | 0.06 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr2_+_9887427 | 0.03 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr10_-_10956700 | 0.03 |
ENSMUST00000105560.2
|
Grm1
|
glutamate receptor, metabotropic 1 |
| chr18_+_37637317 | 0.02 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr17_-_37334091 | 0.02 |
ENSMUST00000167275.3
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr15_+_8997480 | 0.02 |
ENSMUST00000227191.3
|
Ranbp3l
|
RAN binding protein 3-like |
| chr6_-_3494587 | 0.02 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr4_-_82768958 | 0.01 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr19_+_8931187 | 0.01 |
ENSMUST00000096239.7
|
Tut1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
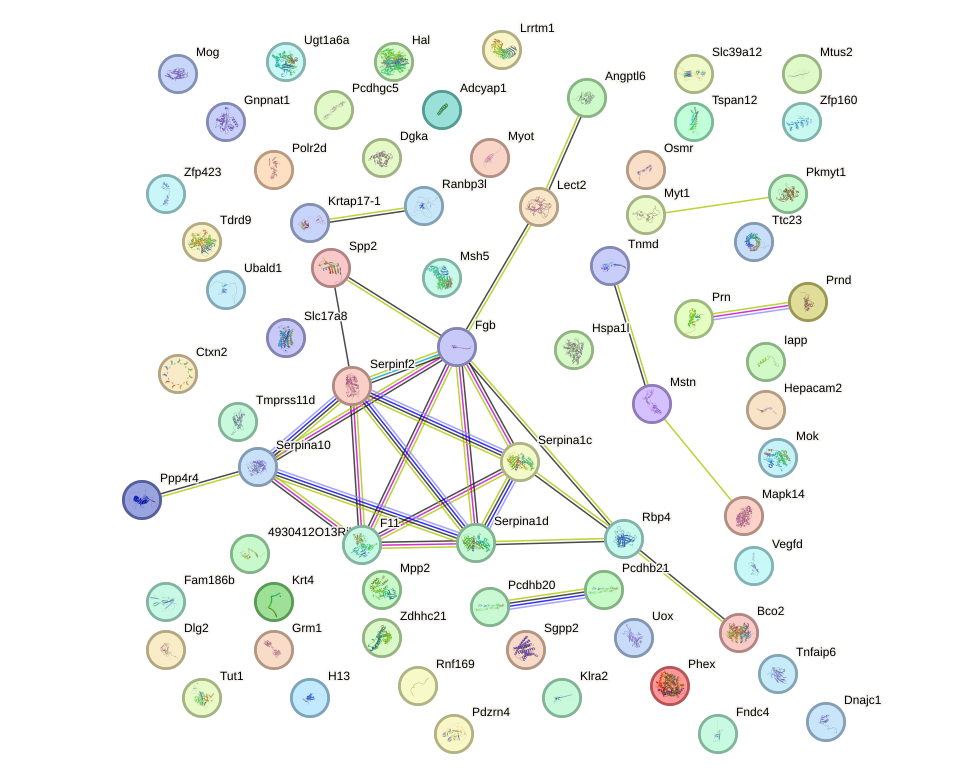
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxd3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.6 | 2.5 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.5 | 1.6 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.4 | 2.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.4 | 1.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.3 | 1.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.3 | 1.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 2.9 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.9 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 3.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.9 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.9 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.5 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.5 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 1.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.6 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.3 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.7 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.1 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.0 | 5.5 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 6.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.0 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) |
| 0.0 | 0.2 | GO:2001269 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001267) positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 1.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.5 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 1.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 1.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 2.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.6 | 4.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.5 | 1.6 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitric oxide binding(GO:0070026) |
| 0.5 | 2.5 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.3 | 3.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 0.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.6 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 1.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 1.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 11.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.3 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 5.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 3.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |