Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Foxn1
Z-value: 0.36
Transcription factors associated with Foxn1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxn1
|
ENSMUSG00000002057.5 | forkhead box N1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxn1 | mm39_v1_chr11_-_78277384_78277433 | -0.02 | 9.0e-01 | Click! |
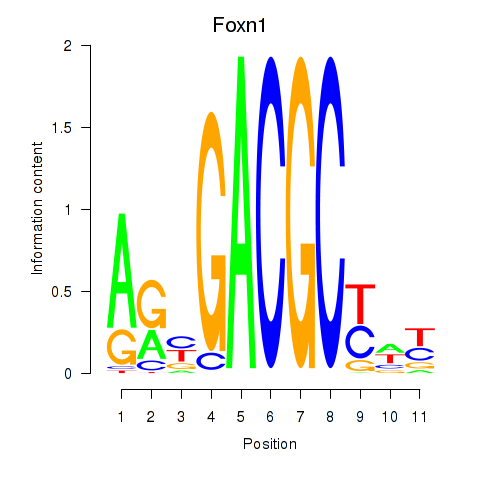
Activity profile of Foxn1 motif
Sorted Z-values of Foxn1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_30721726 | 0.93 |
ENSMUST00000041231.14
|
Psme4
|
proteasome (prosome, macropain) activator subunit 4 |
| chr2_-_125701059 | 0.58 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr9_-_70564403 | 0.54 |
ENSMUST00000213380.2
ENSMUST00000049031.6 |
Mindy2
|
MINDY lysine 48 deubiquitinase 2 |
| chr13_+_81931196 | 0.50 |
ENSMUST00000022009.10
ENSMUST00000223793.2 |
Cetn3
|
centrin 3 |
| chr7_-_81479179 | 0.50 |
ENSMUST00000026093.9
|
Btbd1
|
BTB (POZ) domain containing 1 |
| chr9_+_72892850 | 0.49 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr13_+_81931642 | 0.49 |
ENSMUST00000224574.2
|
Cetn3
|
centrin 3 |
| chr2_-_125700905 | 0.49 |
ENSMUST00000110462.8
|
Cops2
|
COP9 signalosome subunit 2 |
| chr14_-_76348179 | 0.48 |
ENSMUST00000022585.5
|
Gpalpp1
|
GPALPP motifs containing 1 |
| chr9_+_72892693 | 0.48 |
ENSMUST00000037977.15
|
Ccpg1
|
cell cycle progression 1 |
| chr4_+_116414855 | 0.47 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr7_-_133378410 | 0.46 |
ENSMUST00000130182.2
ENSMUST00000106139.8 |
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr19_+_36811615 | 0.46 |
ENSMUST00000025729.12
|
Tnks2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr8_-_84059048 | 0.45 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr19_+_46140942 | 0.44 |
ENSMUST00000026254.14
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr19_+_53933271 | 0.43 |
ENSMUST00000025932.9
|
Shoc2
|
Shoc2, leucine rich repeat scaffold protein |
| chr4_+_116415251 | 0.42 |
ENSMUST00000106475.2
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr7_-_133378468 | 0.42 |
ENSMUST00000033290.12
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr4_+_40722911 | 0.41 |
ENSMUST00000164233.8
ENSMUST00000137246.8 ENSMUST00000125442.8 |
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr1_-_161704224 | 0.39 |
ENSMUST00000048377.11
|
Suco
|
SUN domain containing ossification factor |
| chrX_-_110372843 | 0.39 |
ENSMUST00000096348.10
ENSMUST00000113428.9 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr9_-_59393893 | 0.39 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr13_-_64422775 | 0.37 |
ENSMUST00000221634.2
ENSMUST00000039318.16 |
Cdc14b
|
CDC14 cell division cycle 14B |
| chr19_+_36325683 | 0.37 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr10_+_88215079 | 0.37 |
ENSMUST00000130301.8
ENSMUST00000020251.10 |
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr12_+_105750952 | 0.36 |
ENSMUST00000109901.9
ENSMUST00000168186.8 ENSMUST00000163473.8 ENSMUST00000170540.8 ENSMUST00000166735.8 ENSMUST00000170002.8 |
Papola
|
poly (A) polymerase alpha |
| chr5_-_102217770 | 0.34 |
ENSMUST00000053177.14
ENSMUST00000174698.2 |
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr4_-_121072264 | 0.34 |
ENSMUST00000056635.13
|
Rlf
|
rearranged L-myc fusion sequence |
| chr11_+_93886906 | 0.33 |
ENSMUST00000041956.14
|
Spag9
|
sperm associated antigen 9 |
| chr8_+_61446221 | 0.32 |
ENSMUST00000120689.8
ENSMUST00000034065.14 ENSMUST00000211256.2 ENSMUST00000211672.2 |
Nek1
|
NIMA (never in mitosis gene a)-related expressed kinase 1 |
| chr19_-_14575395 | 0.32 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr14_+_99337311 | 0.32 |
ENSMUST00000022650.9
|
Pibf1
|
progesterone immunomodulatory binding factor 1 |
| chr2_+_18069375 | 0.31 |
ENSMUST00000114671.8
ENSMUST00000114680.9 |
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chrX_-_110373151 | 0.29 |
ENSMUST00000123213.8
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr2_-_163486998 | 0.29 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr11_-_104441218 | 0.29 |
ENSMUST00000106962.9
ENSMUST00000106961.2 ENSMUST00000093923.9 |
Cdc27
|
cell division cycle 27 |
| chr19_-_53933052 | 0.29 |
ENSMUST00000135402.4
|
Bbip1
|
BBSome interacting protein 1 |
| chr3_+_102927901 | 0.28 |
ENSMUST00000198180.5
ENSMUST00000197827.5 ENSMUST00000199240.5 ENSMUST00000199420.5 ENSMUST00000199571.5 ENSMUST00000197488.5 |
Csde1
|
cold shock domain containing E1, RNA binding |
| chr4_+_40722461 | 0.27 |
ENSMUST00000030118.10
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr8_-_94738748 | 0.26 |
ENSMUST00000143265.2
|
Amfr
|
autocrine motility factor receptor |
| chr13_+_38388904 | 0.25 |
ENSMUST00000091641.13
ENSMUST00000178564.2 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr3_+_14643669 | 0.25 |
ENSMUST00000029069.13
ENSMUST00000165922.3 |
E2f5
|
E2F transcription factor 5 |
| chr14_+_75521783 | 0.25 |
ENSMUST00000022577.6
ENSMUST00000227049.2 |
Zc3h13
|
zinc finger CCCH type containing 13 |
| chr8_+_27513839 | 0.25 |
ENSMUST00000209563.2
ENSMUST00000209520.2 |
Erlin2
|
ER lipid raft associated 2 |
| chrX_-_110372800 | 0.24 |
ENSMUST00000137712.9
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr14_-_99337137 | 0.24 |
ENSMUST00000042471.11
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr7_+_16043502 | 0.23 |
ENSMUST00000002152.13
|
Bbc3
|
BCL2 binding component 3 |
| chr9_+_72892786 | 0.23 |
ENSMUST00000156879.8
|
Ccpg1
|
cell cycle progression 1 |
| chr19_-_53932867 | 0.23 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr9_+_70114623 | 0.22 |
ENSMUST00000034745.9
|
Myo1e
|
myosin IE |
| chrX_-_110372733 | 0.22 |
ENSMUST00000065976.12
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr10_+_19810037 | 0.19 |
ENSMUST00000095806.10
ENSMUST00000120259.8 |
Map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr8_+_27513819 | 0.19 |
ENSMUST00000033873.9
ENSMUST00000211043.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr13_-_77283534 | 0.18 |
ENSMUST00000159462.3
ENSMUST00000151524.9 |
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr10_+_88214989 | 0.18 |
ENSMUST00000127615.8
|
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr9_-_106076389 | 0.18 |
ENSMUST00000140761.9
|
Ppm1m
|
protein phosphatase 1M |
| chr10_-_61288437 | 0.17 |
ENSMUST00000167087.2
ENSMUST00000020288.15 |
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr13_+_77283632 | 0.17 |
ENSMUST00000168779.3
ENSMUST00000225605.2 |
2210408I21Rik
|
RIKEN cDNA 2210408I21 gene |
| chr11_-_53525520 | 0.17 |
ENSMUST00000020650.2
|
Il13
|
interleukin 13 |
| chr13_-_64422693 | 0.16 |
ENSMUST00000109770.2
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr9_-_45896075 | 0.16 |
ENSMUST00000217636.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr8_+_27513289 | 0.16 |
ENSMUST00000209795.2
ENSMUST00000209976.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr17_+_47999916 | 0.15 |
ENSMUST00000156118.8
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr11_+_67345895 | 0.15 |
ENSMUST00000108681.9
|
Gas7
|
growth arrest specific 7 |
| chr9_+_69896748 | 0.15 |
ENSMUST00000034754.12
ENSMUST00000085393.13 ENSMUST00000117450.8 |
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr4_+_59003121 | 0.15 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr17_-_65920481 | 0.14 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr9_-_45818196 | 0.13 |
ENSMUST00000160699.9
|
Rnf214
|
ring finger protein 214 |
| chr18_+_33072194 | 0.13 |
ENSMUST00000042868.6
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr3_-_148696155 | 0.13 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr2_+_153334710 | 0.12 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr3_-_115800989 | 0.12 |
ENSMUST00000067485.4
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr16_+_32477722 | 0.12 |
ENSMUST00000238891.2
|
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr9_-_45818134 | 0.12 |
ENSMUST00000161203.8
ENSMUST00000058720.13 |
Rnf214
|
ring finger protein 214 |
| chr2_+_181139016 | 0.11 |
ENSMUST00000108799.10
|
Tpd52l2
|
tumor protein D52-like 2 |
| chr3_+_115801106 | 0.11 |
ENSMUST00000029575.12
ENSMUST00000106501.8 |
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr17_+_87270707 | 0.11 |
ENSMUST00000139344.2
|
Rhoq
|
ras homolog family member Q |
| chr11_-_50101592 | 0.11 |
ENSMUST00000143379.2
ENSMUST00000015981.12 ENSMUST00000102774.11 |
Sqstm1
|
sequestosome 1 |
| chr10_-_127098932 | 0.10 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr7_+_100021425 | 0.10 |
ENSMUST00000098259.11
ENSMUST00000051777.15 |
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr2_+_181138958 | 0.10 |
ENSMUST00000149163.8
ENSMUST00000000844.15 ENSMUST00000184849.8 ENSMUST00000108800.8 ENSMUST00000069712.9 |
Tpd52l2
|
tumor protein D52-like 2 |
| chr8_-_26087552 | 0.10 |
ENSMUST00000210234.2
ENSMUST00000211422.2 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr8_-_94739469 | 0.10 |
ENSMUST00000053766.14
|
Amfr
|
autocrine motility factor receptor |
| chr2_+_109848224 | 0.10 |
ENSMUST00000150183.9
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr6_-_134769558 | 0.10 |
ENSMUST00000100857.10
|
Dusp16
|
dual specificity phosphatase 16 |
| chr6_+_38410848 | 0.10 |
ENSMUST00000160583.8
|
Ubn2
|
ubinuclein 2 |
| chr9_-_45817666 | 0.10 |
ENSMUST00000161187.8
|
Rnf214
|
ring finger protein 214 |
| chr8_-_112603292 | 0.10 |
ENSMUST00000034431.3
|
Tmem170
|
transmembrane protein 170 |
| chr13_-_103911092 | 0.09 |
ENSMUST00000074616.7
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr17_+_26780453 | 0.09 |
ENSMUST00000167662.8
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr11_-_100628979 | 0.09 |
ENSMUST00000155500.2
ENSMUST00000107364.8 ENSMUST00000019317.12 |
Rab5c
|
RAB5C, member RAS oncogene family |
| chr17_+_26780486 | 0.09 |
ENSMUST00000001619.16
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr7_+_133378586 | 0.09 |
ENSMUST00000065359.12
ENSMUST00000209511.2 ENSMUST00000151031.2 ENSMUST00000121560.2 |
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chr8_-_26087475 | 0.08 |
ENSMUST00000210810.2
ENSMUST00000210616.2 ENSMUST00000079160.8 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr10_+_107107558 | 0.08 |
ENSMUST00000105280.5
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr17_-_29456750 | 0.08 |
ENSMUST00000137727.3
|
Cpne5
|
copine V |
| chr17_-_80369762 | 0.08 |
ENSMUST00000061331.14
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr9_-_72892617 | 0.07 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr5_-_46013838 | 0.07 |
ENSMUST00000087164.10
ENSMUST00000121573.8 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr7_+_126447080 | 0.07 |
ENSMUST00000147257.2
ENSMUST00000139174.2 |
Doc2a
|
double C2, alpha |
| chr17_-_25652750 | 0.07 |
ENSMUST00000159610.8
ENSMUST00000159048.8 ENSMUST00000078496.12 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr5_+_98477157 | 0.07 |
ENSMUST00000080333.8
|
Cfap299
|
cilia and flagella associated protein 299 |
| chr2_+_109848151 | 0.06 |
ENSMUST00000028580.12
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr4_-_148021159 | 0.06 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr9_+_45818250 | 0.06 |
ENSMUST00000216672.2
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr10_+_107107477 | 0.05 |
ENSMUST00000020057.16
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr9_-_45896663 | 0.05 |
ENSMUST00000214179.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr9_-_45896110 | 0.05 |
ENSMUST00000215060.2
ENSMUST00000213853.2 ENSMUST00000216334.2 |
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr7_-_109380745 | 0.05 |
ENSMUST00000207400.2
ENSMUST00000033331.7 |
Nrip3
|
nuclear receptor interacting protein 3 |
| chr6_-_39702127 | 0.05 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr9_+_21504018 | 0.05 |
ENSMUST00000062125.11
|
Timm29
|
translocase of inner mitochondrial membrane 29 |
| chr9_+_45817795 | 0.05 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr14_-_30973164 | 0.05 |
ENSMUST00000226565.2
ENSMUST00000022459.5 |
Phf7
|
PHD finger protein 7 |
| chr1_+_134343107 | 0.05 |
ENSMUST00000027727.15
|
Adipor1
|
adiponectin receptor 1 |
| chr2_-_174305856 | 0.05 |
ENSMUST00000016396.8
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr15_-_100952206 | 0.04 |
ENSMUST00000178140.2
|
Fignl2
|
fidgetin-like 2 |
| chr6_-_85490568 | 0.04 |
ENSMUST00000095759.5
|
Egr4
|
early growth response 4 |
| chr14_+_111912529 | 0.03 |
ENSMUST00000042767.9
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr6_+_149310471 | 0.03 |
ENSMUST00000086829.11
ENSMUST00000111513.9 |
Bicd1
|
BICD cargo adaptor 1 |
| chr6_-_39702381 | 0.02 |
ENSMUST00000002487.15
|
Braf
|
Braf transforming gene |
| chr8_+_107662352 | 0.02 |
ENSMUST00000212524.2
ENSMUST00000047425.5 |
Sntb2
|
syntrophin, basic 2 |
| chr5_+_110692162 | 0.02 |
ENSMUST00000040001.14
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr1_+_134343152 | 0.02 |
ENSMUST00000112237.2
|
Adipor1
|
adiponectin receptor 1 |
| chr13_-_55979191 | 0.02 |
ENSMUST00000021968.7
|
Pitx1
|
paired-like homeodomain transcription factor 1 |
| chr1_+_127796508 | 0.02 |
ENSMUST00000037649.6
ENSMUST00000212506.2 |
Rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr19_-_36896999 | 0.02 |
ENSMUST00000238948.2
ENSMUST00000057337.9 |
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr13_-_32522548 | 0.02 |
ENSMUST00000041859.9
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr17_-_80369626 | 0.01 |
ENSMUST00000184635.8
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr14_-_57983511 | 0.01 |
ENSMUST00000173990.8
ENSMUST00000022531.14 |
Lats2
|
large tumor suppressor 2 |
| chr1_+_65965670 | 0.01 |
ENSMUST00000027082.11
ENSMUST00000114027.3 |
Crygf
|
crystallin, gamma F |
| chr2_-_57014015 | 0.01 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr2_+_131052421 | 0.00 |
ENSMUST00000110206.2
|
Ap5s1
|
adaptor-related protein 5 complex, sigma 1 subunit |
| chr13_+_55612050 | 0.00 |
ENSMUST00000046533.9
|
Prr7
|
proline rich 7 (synaptic) |
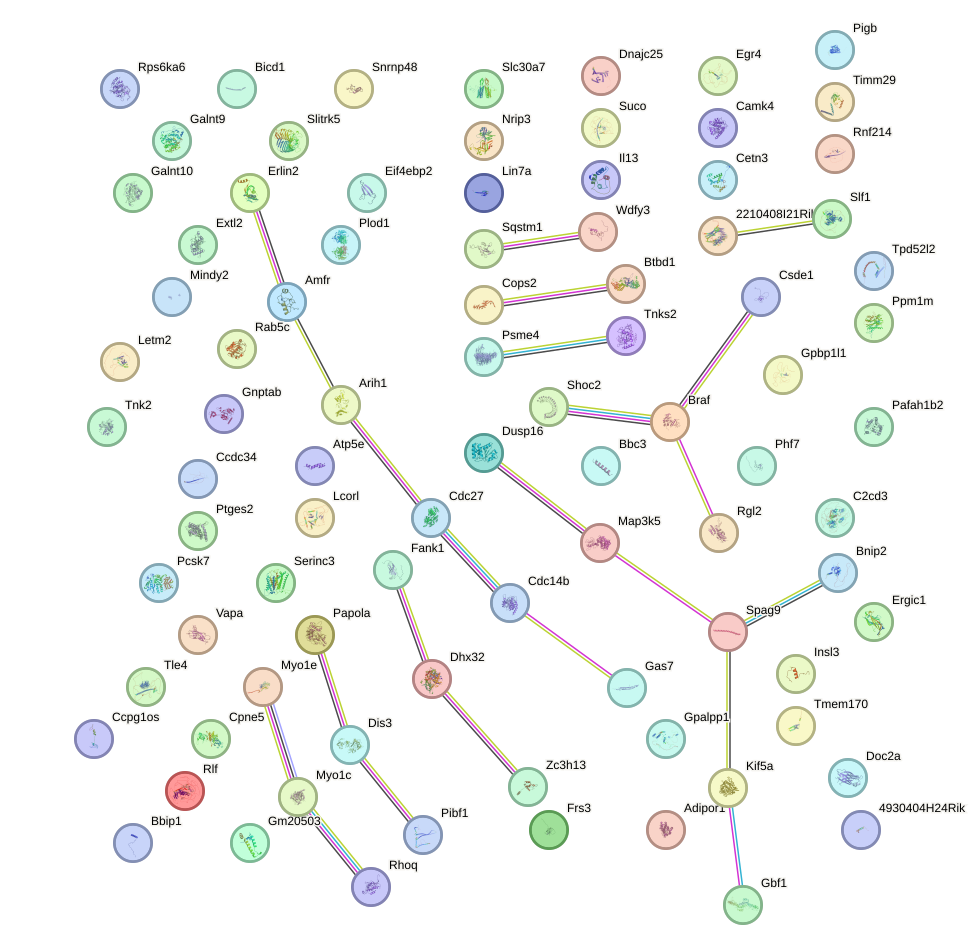
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxn1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.2 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.5 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 1.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.2 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 1.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.6 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.2 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.2 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0061511 | neural plate axis specification(GO:0021997) centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.0 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.5 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 1.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |