Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
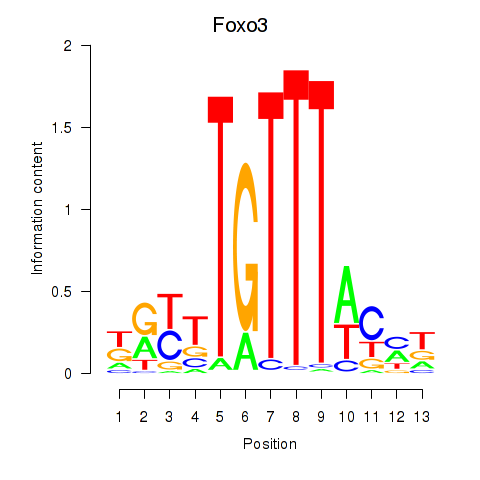
Results for Foxo3
Z-value: 0.70
Transcription factors associated with Foxo3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo3
|
ENSMUSG00000048756.12 | forkhead box O3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo3 | mm39_v1_chr10_-_42152684_42152759 | -0.80 | 3.4e-09 | Click! |
Activity profile of Foxo3 motif
Sorted Z-values of Foxo3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_51333816 | 7.69 |
ENSMUST00000169895.3
|
Rnase4
|
ribonuclease, RNase A family 4 |
| chr14_-_52150804 | 6.63 |
ENSMUST00000004673.15
ENSMUST00000111632.5 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr14_-_52151026 | 5.61 |
ENSMUST00000228164.2
|
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr1_-_180023518 | 5.53 |
ENSMUST00000162769.8
ENSMUST00000161379.2 ENSMUST00000027766.13 ENSMUST00000161814.8 |
Coq8a
|
coenzyme Q8A |
| chr1_-_180023467 | 5.52 |
ENSMUST00000161746.2
ENSMUST00000160879.7 |
Coq8a
|
coenzyme Q8A |
| chr15_-_78352801 | 5.33 |
ENSMUST00000229124.2
ENSMUST00000230226.2 ENSMUST00000017086.5 |
Tmprss6
|
transmembrane serine protease 6 |
| chr11_+_101258368 | 4.49 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr4_-_49549489 | 4.08 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr9_-_71070506 | 3.95 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr8_+_105996419 | 3.33 |
ENSMUST00000036127.9
ENSMUST00000163734.9 |
Hsf4
|
heat shock transcription factor 4 |
| chr1_-_140111138 | 3.06 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 2.94 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr2_+_155360015 | 2.83 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr3_+_85946145 | 2.71 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr2_+_155359868 | 2.66 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr1_+_172525613 | 2.64 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr4_+_105014536 | 2.50 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr11_-_86884507 | 2.45 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr19_-_7780025 | 2.31 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr9_+_114560235 | 2.27 |
ENSMUST00000035007.10
|
Cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr8_-_45835234 | 2.16 |
ENSMUST00000093526.13
|
Fam149a
|
family with sequence similarity 149, member A |
| chr1_+_171052623 | 2.02 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr2_+_76505619 | 1.97 |
ENSMUST00000111920.2
|
Plekha3
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 3 |
| chr11_-_60243695 | 1.95 |
ENSMUST00000095254.12
ENSMUST00000102683.11 ENSMUST00000093048.13 ENSMUST00000093046.13 ENSMUST00000064019.15 ENSMUST00000102682.5 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr8_-_64659004 | 1.92 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr3_+_63203516 | 1.88 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr8_-_26275182 | 1.85 |
ENSMUST00000038498.10
|
Bag4
|
BCL2-associated athanogene 4 |
| chr2_+_4722956 | 1.82 |
ENSMUST00000056914.7
|
Bend7
|
BEN domain containing 7 |
| chr5_-_151113619 | 1.74 |
ENSMUST00000062015.15
ENSMUST00000110483.9 |
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr14_+_30601157 | 1.73 |
ENSMUST00000040715.8
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr5_+_65288418 | 1.73 |
ENSMUST00000101191.10
ENSMUST00000204348.3 |
Klhl5
|
kelch-like 5 |
| chr4_-_155430153 | 1.65 |
ENSMUST00000103178.11
|
Prkcz
|
protein kinase C, zeta |
| chr3_+_107137924 | 1.63 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr1_+_75119419 | 1.62 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr17_+_25059079 | 1.57 |
ENSMUST00000164251.8
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr19_+_5927821 | 1.53 |
ENSMUST00000145200.8
ENSMUST00000025732.14 ENSMUST00000125114.8 ENSMUST00000155697.8 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr19_+_34268071 | 1.53 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr19_-_34856853 | 1.43 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chrX_-_47297746 | 1.40 |
ENSMUST00000088935.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr18_-_75094323 | 1.38 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr2_-_51862941 | 1.38 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr3_+_63203235 | 1.37 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr11_-_68277631 | 1.36 |
ENSMUST00000021284.4
|
Ntn1
|
netrin 1 |
| chr19_+_5927876 | 1.32 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr2_-_104324035 | 1.31 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr1_+_75119472 | 1.21 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr13_+_60749995 | 1.17 |
ENSMUST00000044083.9
|
Dapk1
|
death associated protein kinase 1 |
| chr12_-_25147139 | 1.11 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr7_-_65020955 | 1.08 |
ENSMUST00000102592.10
|
Tjp1
|
tight junction protein 1 |
| chr2_+_3514071 | 1.08 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr2_-_91540864 | 1.04 |
ENSMUST00000028678.9
ENSMUST00000076803.12 |
Atg13
|
autophagy related 13 |
| chr2_+_91541245 | 1.00 |
ENSMUST00000142692.2
ENSMUST00000090608.6 |
Harbi1
|
harbinger transposase derived 1 |
| chr17_+_43700327 | 0.99 |
ENSMUST00000113599.2
ENSMUST00000224278.2 ENSMUST00000225466.2 |
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr7_-_70010341 | 0.92 |
ENSMUST00000032768.15
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_-_83808717 | 0.90 |
ENSMUST00000058383.9
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr11_-_68277799 | 0.86 |
ENSMUST00000135141.2
|
Ntn1
|
netrin 1 |
| chr9_-_48747232 | 0.86 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr18_-_39622829 | 0.85 |
ENSMUST00000025300.13
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr14_+_45457168 | 0.83 |
ENSMUST00000227086.2
ENSMUST00000147957.2 |
Gpr137c
|
G protein-coupled receptor 137C |
| chr6_+_14901343 | 0.82 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr18_-_39622932 | 0.79 |
ENSMUST00000152853.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr10_+_88036947 | 0.78 |
ENSMUST00000020248.16
ENSMUST00000182183.8 ENSMUST00000171151.9 ENSMUST00000182619.2 |
Washc3
|
WASH complex subunit 3 |
| chr4_-_119272690 | 0.75 |
ENSMUST00000238287.2
ENSMUST00000238759.2 ENSMUST00000063642.10 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr4_-_91264670 | 0.75 |
ENSMUST00000107109.9
ENSMUST00000107111.9 ENSMUST00000107120.8 |
Elavl2
|
ELAV like RNA binding protein 1 |
| chr2_+_91541197 | 0.74 |
ENSMUST00000128140.2
ENSMUST00000140183.2 |
Harbi1
|
harbinger transposase derived 1 |
| chr2_-_51863203 | 0.72 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr4_+_135455427 | 0.71 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr11_+_96024612 | 0.69 |
ENSMUST00000167258.8
|
Ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr18_-_39622295 | 0.67 |
ENSMUST00000131885.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr14_-_121935829 | 0.66 |
ENSMUST00000040700.9
ENSMUST00000212181.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr1_-_20854490 | 0.66 |
ENSMUST00000039046.10
|
Il17f
|
interleukin 17F |
| chr2_-_27365633 | 0.65 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr8_-_41469786 | 0.65 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr4_-_91264727 | 0.64 |
ENSMUST00000107124.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr6_+_14901439 | 0.63 |
ENSMUST00000128567.8
|
Foxp2
|
forkhead box P2 |
| chr7_+_107264518 | 0.57 |
ENSMUST00000239294.2
|
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr8_-_13939964 | 0.57 |
ENSMUST00000209371.2
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr10_+_103203552 | 0.51 |
ENSMUST00000179636.3
ENSMUST00000217905.2 ENSMUST00000074204.12 |
Slc6a15
|
solute carrier family 6 (neurotransmitter transporter), member 15 |
| chr7_-_65020655 | 0.51 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr4_-_119272640 | 0.50 |
ENSMUST00000238293.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr19_+_56414114 | 0.50 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr11_+_60244132 | 0.49 |
ENSMUST00000070805.13
ENSMUST00000094140.9 ENSMUST00000108723.9 ENSMUST00000108722.5 |
Drc3
|
dynein regulatory complex subunit 3 |
| chr10_+_128540049 | 0.48 |
ENSMUST00000217836.2
|
Pmel
|
premelanosome protein |
| chr12_-_85317359 | 0.47 |
ENSMUST00000166821.8
ENSMUST00000019378.8 ENSMUST00000220854.2 |
Mlh3
|
mutL homolog 3 |
| chr11_-_101676076 | 0.47 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr11_+_82802079 | 0.45 |
ENSMUST00000018989.14
ENSMUST00000164945.3 |
Unc45b
|
unc-45 myosin chaperone B |
| chr7_+_141056305 | 0.43 |
ENSMUST00000117634.2
|
Tspan4
|
tetraspanin 4 |
| chr4_-_119272667 | 0.42 |
ENSMUST00000238609.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr3_-_145355725 | 0.41 |
ENSMUST00000029846.5
|
Ccn1
|
cellular communication network factor 1 |
| chr2_+_164328375 | 0.40 |
ENSMUST00000069385.15
ENSMUST00000143690.8 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_-_100244866 | 0.39 |
ENSMUST00000173630.8
|
Hap1
|
huntingtin-associated protein 1 |
| chr17_+_31783708 | 0.39 |
ENSMUST00000097352.11
ENSMUST00000237248.2 ENSMUST00000235869.2 ENSMUST00000175806.9 |
Pknox1
|
Pbx/knotted 1 homeobox |
| chr10_-_5019044 | 0.39 |
ENSMUST00000095899.5
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr4_+_130001349 | 0.39 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr9_+_40092216 | 0.37 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr15_+_102875229 | 0.37 |
ENSMUST00000001699.8
|
Hoxc10
|
homeobox C10 |
| chr10_+_102348076 | 0.37 |
ENSMUST00000219445.2
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr11_-_99313078 | 0.36 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr3_+_53396120 | 0.35 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr13_+_83720484 | 0.34 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr15_-_77129706 | 0.32 |
ENSMUST00000228361.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr18_-_66155651 | 0.30 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr9_+_65268304 | 0.30 |
ENSMUST00000147185.3
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr1_+_66360865 | 0.29 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr19_-_27988534 | 0.28 |
ENSMUST00000174850.8
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr8_+_72021510 | 0.28 |
ENSMUST00000212889.2
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr19_+_25649767 | 0.26 |
ENSMUST00000053068.7
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chr11_+_3282424 | 0.26 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_+_17672117 | 0.25 |
ENSMUST00000088476.4
|
Pi15
|
peptidase inhibitor 15 |
| chr8_+_72021567 | 0.24 |
ENSMUST00000034267.5
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr2_+_164328763 | 0.24 |
ENSMUST00000109349.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_+_127696548 | 0.23 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr1_-_58735106 | 0.23 |
ENSMUST00000055313.14
ENSMUST00000188772.7 |
Flacc1
|
flagellum associated containing coiled-coil domains 1 |
| chrX_+_7744535 | 0.22 |
ENSMUST00000033495.15
|
Pim2
|
proviral integration site 2 |
| chr3_-_146475974 | 0.20 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr16_-_34083315 | 0.20 |
ENSMUST00000114953.8
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr4_-_134642286 | 0.20 |
ENSMUST00000105863.2
ENSMUST00000030626.12 |
Tmem50a
|
transmembrane protein 50A |
| chr10_+_25308466 | 0.19 |
ENSMUST00000219224.2
ENSMUST00000219166.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr13_+_108180957 | 0.19 |
ENSMUST00000095458.6
ENSMUST00000223808.2 ENSMUST00000226042.2 ENSMUST00000225972.2 |
Smim15
|
small integral membrane protein 15 |
| chr15_+_25752963 | 0.18 |
ENSMUST00000022882.12
ENSMUST00000135173.8 |
Myo10
|
myosin X |
| chr13_+_83720457 | 0.16 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr3_+_5283606 | 0.15 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr1_+_36550948 | 0.15 |
ENSMUST00000001166.14
ENSMUST00000097776.4 |
Cnnm3
|
cyclin M3 |
| chr2_+_154855350 | 0.13 |
ENSMUST00000148402.8
|
a
|
nonagouti |
| chr14_-_52011035 | 0.12 |
ENSMUST00000073860.6
|
Ang4
|
angiogenin, ribonuclease A family, member 4 |
| chr2_-_7086066 | 0.12 |
ENSMUST00000183209.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr7_-_119461027 | 0.11 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr1_-_172958803 | 0.11 |
ENSMUST00000073663.3
|
Olfr1408
|
olfactory receptor 1408 |
| chr3_+_5283577 | 0.11 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr10_+_112107026 | 0.11 |
ENSMUST00000219301.2
ENSMUST00000092175.4 |
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr2_-_51039112 | 0.11 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr6_+_55813862 | 0.10 |
ENSMUST00000044729.7
|
Itprid1
|
ITPR interacting domain containing 1 |
| chr12_-_31549538 | 0.09 |
ENSMUST00000064240.14
ENSMUST00000185739.8 ENSMUST00000188326.3 ENSMUST00000101499.10 ENSMUST00000085487.12 |
Cbll1
|
Casitas B-lineage lymphoma-like 1 |
| chr16_+_93480061 | 0.08 |
ENSMUST00000039620.7
|
Cbr3
|
carbonyl reductase 3 |
| chr11_+_19874403 | 0.08 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr6_+_34840151 | 0.07 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr9_+_78020554 | 0.06 |
ENSMUST00000009972.6
ENSMUST00000117330.8 ENSMUST00000044551.8 |
Cilk1
|
ciliogenesis associated kinase 1 |
| chr1_-_163231196 | 0.04 |
ENSMUST00000045138.6
|
Gorab
|
golgin, RAB6-interacting |
| chr14_-_7750938 | 0.03 |
ENSMUST00000223740.3
ENSMUST00000224049.3 ENSMUST00000224672.3 ENSMUST00000224752.3 ENSMUST00000223695.3 ENSMUST00000224333.3 ENSMUST00000224952.3 |
Slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr6_-_69741999 | 0.03 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr7_-_109215754 | 0.03 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chr2_-_85675827 | 0.02 |
ENSMUST00000213515.2
|
Olfr1019
|
olfactory receptor 1019 |
| chr7_+_29991366 | 0.02 |
ENSMUST00000144508.2
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr14_-_57371041 | 0.02 |
ENSMUST00000039380.9
|
Gjb6
|
gap junction protein, beta 6 |
| chr19_-_19088543 | 0.02 |
ENSMUST00000112832.8
|
Rorb
|
RAR-related orphan receptor beta |
| chr1_+_63655127 | 0.01 |
ENSMUST00000226288.2
|
Gm39653
|
predicted gene, 39653 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxo3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.2 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.8 | 5.5 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 1.0 | 3.9 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.7 | 2.1 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.7 | 4.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.7 | 6.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.6 | 1.8 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.5 | 1.5 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.5 | 2.0 | GO:0046340 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
| 0.5 | 2.5 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.5 | 4.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.5 | 3.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.5 | 1.4 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.5 | 11.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.4 | 1.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.3 | 1.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 2.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 2.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 5.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.7 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 0.9 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 | 1.7 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.2 | 2.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 0.4 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 1.6 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 2.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 3.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.7 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.1 | 0.5 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 2.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.5 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.9 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 1.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 1.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.4 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.1 | 1.0 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.1 | 1.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.5 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.5 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.4 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.0 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.6 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 7.0 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 2.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.1 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.2 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 1.2 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 1.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 0.5 | GO:0005712 | chiasma(GO:0005712) |
| 0.2 | 1.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 1.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 4.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.4 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 2.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 3.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.1 | 5.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.9 | 6.0 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.8 | 3.9 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.6 | 2.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.5 | 2.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 4.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 2.3 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.4 | 1.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 2.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 1.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 11.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 0.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 2.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 2.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 0.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 2.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 6.6 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 3.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 3.3 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.7 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 13.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 5.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 2.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 6.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 2.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 3.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 4.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 4.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 2.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |