Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
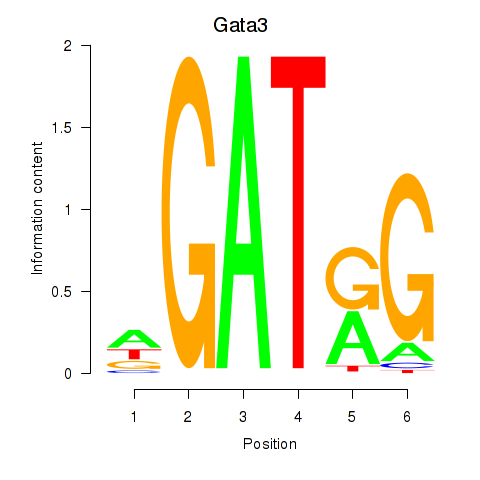
Results for Gata3
Z-value: 1.95
Transcription factors associated with Gata3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata3
|
ENSMUSG00000015619.11 | GATA binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata3 | mm39_v1_chr2_-_9883391_9883418 | -0.45 | 5.9e-03 | Click! |
Activity profile of Gata3 motif
Sorted Z-values of Gata3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_97066937 | 9.70 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr4_-_60455331 | 9.45 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr11_+_32226400 | 6.30 |
ENSMUST00000020531.9
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr2_+_155359868 | 5.99 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr2_+_155360015 | 5.40 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr11_+_32226893 | 4.55 |
ENSMUST00000145569.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr7_-_103492361 | 4.16 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr7_+_26534730 | 4.12 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr11_+_95715295 | 3.97 |
ENSMUST00000150134.2
ENSMUST00000054173.4 |
Phospho1
|
phosphatase, orphan 1 |
| chr7_-_103502404 | 3.92 |
ENSMUST00000033229.5
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr17_+_85335775 | 3.83 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr3_-_107876477 | 3.68 |
ENSMUST00000004136.10
|
Gstm3
|
glutathione S-transferase, mu 3 |
| chr2_+_102489558 | 3.38 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr13_+_23930717 | 3.30 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr7_+_26006594 | 3.26 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr11_-_84058292 | 3.23 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr11_-_100418717 | 2.81 |
ENSMUST00000107389.8
ENSMUST00000007131.16 |
Acly
|
ATP citrate lyase |
| chr1_-_75195127 | 2.49 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr19_-_46661321 | 2.34 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr14_+_30930266 | 2.32 |
ENSMUST00000169169.8
|
Tnnc1
|
troponin C, cardiac/slow skeletal |
| chr5_-_4154681 | 2.28 |
ENSMUST00000001507.5
|
Cyp51
|
cytochrome P450, family 51 |
| chr1_+_165596961 | 2.27 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr11_-_100418688 | 2.23 |
ENSMUST00000107385.2
|
Acly
|
ATP citrate lyase |
| chr19_-_46661501 | 2.13 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr2_+_111610658 | 2.05 |
ENSMUST00000054004.2
|
Olfr1302
|
olfactory receptor 1302 |
| chr8_+_70536103 | 2.03 |
ENSMUST00000007738.11
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr15_+_59186876 | 1.92 |
ENSMUST00000022977.14
ENSMUST00000100640.5 |
Sqle
|
squalene epoxidase |
| chr11_-_84719779 | 1.89 |
ENSMUST00000047560.8
|
Dhrs11
|
dehydrogenase/reductase (SDR family) member 11 |
| chr7_-_133310687 | 1.84 |
ENSMUST00000106146.8
|
Uros
|
uroporphyrinogen III synthase |
| chr7_-_133310779 | 1.77 |
ENSMUST00000124759.2
ENSMUST00000106144.8 ENSMUST00000106145.10 |
Uros
|
uroporphyrinogen III synthase |
| chr6_-_124441731 | 1.69 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr18_-_25302064 | 1.64 |
ENSMUST00000115817.3
|
Tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr1_+_165591315 | 1.63 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr9_+_54606832 | 1.61 |
ENSMUST00000070070.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr1_-_75193937 | 1.61 |
ENSMUST00000186971.2
ENSMUST00000188593.2 |
Tuba4a
|
tubulin, alpha 4A |
| chr2_+_109522781 | 1.56 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr4_+_13751297 | 1.55 |
ENSMUST00000105566.9
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr7_+_127344942 | 1.54 |
ENSMUST00000189562.7
ENSMUST00000186116.7 |
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr15_-_101833160 | 1.50 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr9_-_44253630 | 1.48 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr7_+_25358589 | 1.47 |
ENSMUST00000079634.8
ENSMUST00000206561.2 |
Exosc5
|
exosome component 5 |
| chr9_+_102885156 | 1.45 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr11_+_102727122 | 1.43 |
ENSMUST00000021302.15
ENSMUST00000107072.2 |
Higd1b
|
HIG1 domain family, member 1B |
| chr11_+_101207743 | 1.41 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr17_+_37253802 | 1.35 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr7_-_25069128 | 1.34 |
ENSMUST00000167591.8
ENSMUST00000076276.5 |
Cnfn
|
cornifelin |
| chr13_-_76205115 | 1.33 |
ENSMUST00000056130.8
|
Gpr150
|
G protein-coupled receptor 150 |
| chr8_-_76133212 | 1.33 |
ENSMUST00000212864.2
|
Gm10358
|
predicted gene 10358 |
| chr14_-_70864448 | 1.33 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr13_+_23715220 | 1.31 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr3_+_96175970 | 1.28 |
ENSMUST00000098843.3
|
H3c13
|
H3 clustered histone 13 |
| chr15_+_97990431 | 1.26 |
ENSMUST00000229280.2
ENSMUST00000163507.8 ENSMUST00000230445.2 |
Pfkm
|
phosphofructokinase, muscle |
| chr8_+_85696695 | 1.25 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr4_+_117706390 | 1.24 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr4_+_106591080 | 1.22 |
ENSMUST00000047620.3
|
Fam151a
|
family with sequence simliarity 151, member A |
| chr8_+_123829089 | 1.21 |
ENSMUST00000000756.6
|
Rpl13
|
ribosomal protein L13 |
| chrX_+_100492684 | 1.20 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chrX_-_36166529 | 1.18 |
ENSMUST00000057093.8
|
Nkrf
|
NF-kappaB repressing factor |
| chr17_+_35658131 | 1.18 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr15_+_76580925 | 1.17 |
ENSMUST00000023203.6
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr13_+_21906214 | 1.17 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr15_+_74388044 | 1.16 |
ENSMUST00000042035.16
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr11_-_95733235 | 1.16 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr9_-_21913833 | 1.15 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr8_+_85696396 | 1.15 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr8_+_85696453 | 1.15 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr13_-_21685588 | 1.13 |
ENSMUST00000044043.3
|
Cox5b-ps
|
cytochrome c oxidase subunit 5B, pseudogene |
| chr6_-_83030759 | 1.12 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr15_+_76582372 | 1.09 |
ENSMUST00000229140.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr11_+_49170977 | 1.09 |
ENSMUST00000078932.2
|
Olfr1393
|
olfactory receptor 1393 |
| chr8_-_65186565 | 1.09 |
ENSMUST00000141021.2
|
Msmo1
|
methylsterol monoxygenase 1 |
| chr7_+_78432867 | 1.07 |
ENSMUST00000032840.5
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr9_-_107960528 | 1.07 |
ENSMUST00000159372.3
ENSMUST00000160249.8 |
Rnf123
|
ring finger protein 123 |
| chr4_-_43045685 | 1.06 |
ENSMUST00000107956.8
ENSMUST00000107957.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr8_-_85692634 | 1.06 |
ENSMUST00000109738.10
ENSMUST00000065049.15 ENSMUST00000128972.9 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr9_-_21900724 | 1.06 |
ENSMUST00000045726.8
|
Rgl3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chrX_-_8059597 | 1.05 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr13_+_23755551 | 1.05 |
ENSMUST00000079251.8
|
H2bc8
|
H2B clustered histone 8 |
| chr3_-_107992662 | 1.04 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr2_+_121978156 | 1.04 |
ENSMUST00000102476.5
|
B2m
|
beta-2 microglobulin |
| chr11_-_93856783 | 1.04 |
ENSMUST00000021220.10
|
Nme1
|
NME/NM23 nucleoside diphosphate kinase 1 |
| chr11_-_100012384 | 1.04 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chrX_-_8072714 | 1.02 |
ENSMUST00000089403.10
ENSMUST00000077595.12 ENSMUST00000089402.10 ENSMUST00000082320.12 |
Porcn
|
porcupine O-acyltransferase |
| chr9_-_39918243 | 1.02 |
ENSMUST00000073932.4
|
Olfr980
|
olfactory receptor 980 |
| chr7_-_44145830 | 1.01 |
ENSMUST00000118515.9
ENSMUST00000138328.3 ENSMUST00000239015.2 ENSMUST00000118808.9 |
Emc10
|
ER membrane protein complex subunit 10 |
| chr2_-_65397850 | 1.01 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr7_-_99340830 | 1.01 |
ENSMUST00000208713.2
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr7_+_126294527 | 1.01 |
ENSMUST00000130498.2
|
Bola2
|
bolA-like 2 (E. coli) |
| chr18_-_75094323 | 1.01 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chrX_+_100298134 | 1.01 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr17_+_25115461 | 0.99 |
ENSMUST00000024978.7
|
Nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr4_+_56740070 | 0.99 |
ENSMUST00000181745.2
|
Gm26657
|
predicted gene, 26657 |
| chr13_-_23755374 | 0.99 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr15_-_103159892 | 0.99 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr5_-_24652775 | 0.98 |
ENSMUST00000123167.2
ENSMUST00000030799.15 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr14_+_61844899 | 0.98 |
ENSMUST00000225582.2
ENSMUST00000051184.10 |
Kcnrg
|
potassium channel regulator |
| chr3_+_96177010 | 0.96 |
ENSMUST00000051089.4
ENSMUST00000177113.2 |
Gm42743
H2bc18
|
predicted gene 42743 H2B clustered histone 18 |
| chr2_+_30306116 | 0.95 |
ENSMUST00000113601.10
ENSMUST00000113603.10 |
Ptpa
|
protein phosphatase 2 protein activator |
| chr10_+_23846604 | 0.94 |
ENSMUST00000092659.4
|
Taar5
|
trace amine-associated receptor 5 |
| chr12_+_84820024 | 0.94 |
ENSMUST00000021667.7
ENSMUST00000222449.2 ENSMUST00000222982.2 |
Isca2
|
iron-sulfur cluster assembly 2 |
| chr13_-_76166789 | 0.94 |
ENSMUST00000179078.9
ENSMUST00000167271.9 |
Rfesd
|
Rieske (Fe-S) domain containing |
| chr12_+_84820095 | 0.94 |
ENSMUST00000222022.2
|
Isca2
|
iron-sulfur cluster assembly 2 |
| chr15_-_101602734 | 0.94 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr4_+_43631935 | 0.93 |
ENSMUST00000030191.15
|
Npr2
|
natriuretic peptide receptor 2 |
| chr3_-_96128196 | 0.93 |
ENSMUST00000090782.4
|
H2ac20
|
H2A clustered histone 20 |
| chr13_-_3854307 | 0.92 |
ENSMUST00000077698.5
|
Calml3
|
calmodulin-like 3 |
| chr4_+_58285957 | 0.92 |
ENSMUST00000081919.12
ENSMUST00000177951.8 ENSMUST00000098059.10 ENSMUST00000179951.2 ENSMUST00000102893.10 ENSMUST00000084578.12 ENSMUST00000098057.10 |
Musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr9_-_99592116 | 0.92 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr7_+_103578032 | 0.91 |
ENSMUST00000106863.2
|
Olfr631
|
olfactory receptor 631 |
| chr6_-_115228800 | 0.91 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr15_+_83447784 | 0.90 |
ENSMUST00000047419.8
|
Tspo
|
translocator protein |
| chr19_+_5651882 | 0.89 |
ENSMUST00000025864.11
ENSMUST00000236461.2 ENSMUST00000155639.2 |
Rnaseh2c
|
ribonuclease H2, subunit C |
| chr2_-_65397809 | 0.89 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr1_-_75166994 | 0.89 |
ENSMUST00000189820.7
|
Atg9a
|
autophagy related 9A |
| chrX_+_149377416 | 0.89 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr1_-_65142521 | 0.88 |
ENSMUST00000061497.9
|
Cryga
|
crystallin, gamma A |
| chr14_-_36832044 | 0.88 |
ENSMUST00000179488.3
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr7_+_126295114 | 0.87 |
ENSMUST00000106369.2
|
Bola2
|
bolA-like 2 (E. coli) |
| chr19_+_6108240 | 0.87 |
ENSMUST00000237840.2
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr11_-_59029996 | 0.87 |
ENSMUST00000219084.3
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr6_-_124790029 | 0.87 |
ENSMUST00000149610.3
|
Tpi1
|
triosephosphate isomerase 1 |
| chr2_+_90865958 | 0.87 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr4_+_150233362 | 0.87 |
ENSMUST00000059893.8
|
Slc2a7
|
solute carrier family 2 (facilitated glucose transporter), member 7 |
| chrX_+_139357362 | 0.86 |
ENSMUST00000033809.4
|
Prps1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr4_+_102971909 | 0.86 |
ENSMUST00000143417.8
|
Mier1
|
MEIR1 treanscription regulator |
| chr2_+_30306045 | 0.86 |
ENSMUST00000042055.10
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr7_-_126194097 | 0.86 |
ENSMUST00000058429.6
|
Il27
|
interleukin 27 |
| chr15_-_77854988 | 0.85 |
ENSMUST00000100484.6
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr11_-_101315345 | 0.85 |
ENSMUST00000107257.8
ENSMUST00000107259.4 ENSMUST00000107252.9 ENSMUST00000093933.11 |
Gm27029
Ptges3l
|
predicted gene, 27029 prostaglandin E synthase 3 like |
| chr6_-_113354826 | 0.84 |
ENSMUST00000032410.14
|
Tada3
|
transcriptional adaptor 3 |
| chr4_+_123095297 | 0.84 |
ENSMUST00000068262.6
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr13_+_78173013 | 0.84 |
ENSMUST00000175955.4
|
Pou5f2
|
POU domain class 5, transcription factor 2 |
| chr11_+_70104736 | 0.84 |
ENSMUST00000171032.8
|
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr14_-_50536787 | 0.84 |
ENSMUST00000163469.2
|
Olfr733
|
olfactory receptor 733 |
| chr11_+_70410445 | 0.84 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr15_+_7159038 | 0.84 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr1_+_93682621 | 0.83 |
ENSMUST00000027502.16
|
Atg4b
|
autophagy related 4B, cysteine peptidase |
| chr8_-_68270870 | 0.82 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_31274064 | 0.82 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr19_-_11833365 | 0.82 |
ENSMUST00000079875.4
|
Olfr1418
|
olfactory receptor 1418 |
| chr15_-_98779134 | 0.81 |
ENSMUST00000229348.2
ENSMUST00000229876.2 |
Rhebl1
|
Ras homolog enriched in brain like 1 |
| chr7_-_44785480 | 0.81 |
ENSMUST00000211246.2
ENSMUST00000210197.2 |
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr9_-_99592058 | 0.81 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr5_+_31274046 | 0.80 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr4_+_60003438 | 0.80 |
ENSMUST00000107517.8
ENSMUST00000107520.2 |
Mup6
|
major urinary protein 6 |
| chr17_+_37253916 | 0.80 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr18_+_61820982 | 0.80 |
ENSMUST00000025471.4
|
Il17b
|
interleukin 17B |
| chr11_+_87651359 | 0.79 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr5_+_16139909 | 0.79 |
ENSMUST00000196750.2
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr11_-_72026547 | 0.79 |
ENSMUST00000108508.3
ENSMUST00000075258.13 |
Pitpnm3
|
PITPNM family member 3 |
| chr18_-_78640066 | 0.79 |
ENSMUST00000235389.2
ENSMUST00000237674.2 |
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr1_+_166081755 | 0.78 |
ENSMUST00000194964.6
ENSMUST00000192638.6 ENSMUST00000192426.6 ENSMUST00000195557.6 ENSMUST00000192732.6 ENSMUST00000193860.2 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr12_-_8589545 | 0.78 |
ENSMUST00000095863.10
ENSMUST00000165657.3 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr7_+_103628383 | 0.78 |
ENSMUST00000098185.2
|
Olfr635
|
olfactory receptor 635 |
| chr10_+_86893907 | 0.78 |
ENSMUST00000189456.7
ENSMUST00000169849.3 |
1700113H08Rik
|
RIKEN cDNA 1700113H08 gene |
| chr16_+_43184191 | 0.78 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_-_63663344 | 0.77 |
ENSMUST00000239480.2
ENSMUST00000062246.8 |
Tnfsf15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr3_+_94385661 | 0.77 |
ENSMUST00000200342.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr4_-_3973586 | 0.77 |
ENSMUST00000089430.6
|
Gm11808
|
predicted gene 11808 |
| chr2_+_112092271 | 0.77 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr4_-_22488296 | 0.77 |
ENSMUST00000178174.3
|
Pou3f2
|
POU domain, class 3, transcription factor 2 |
| chr11_-_97913420 | 0.77 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr5_+_31212110 | 0.77 |
ENSMUST00000013773.12
ENSMUST00000201838.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr1_+_134343152 | 0.77 |
ENSMUST00000112237.2
|
Adipor1
|
adiponectin receptor 1 |
| chr17_+_37209002 | 0.77 |
ENSMUST00000078438.5
|
Trim31
|
tripartite motif-containing 31 |
| chr9_+_119943916 | 0.77 |
ENSMUST00000135514.2
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr6_+_83055581 | 0.76 |
ENSMUST00000177177.8
ENSMUST00000176089.2 |
Pcgf1
|
polycomb group ring finger 1 |
| chr4_-_63090355 | 0.76 |
ENSMUST00000156618.9
ENSMUST00000030042.3 |
Kif12
|
kinesin family member 12 |
| chr5_-_136596094 | 0.76 |
ENSMUST00000175975.9
ENSMUST00000176216.9 ENSMUST00000176745.8 |
Cux1
|
cut-like homeobox 1 |
| chr4_-_82803384 | 0.76 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr13_+_46655324 | 0.76 |
ENSMUST00000021802.16
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr4_+_129083553 | 0.76 |
ENSMUST00000106054.4
|
Yars
|
tyrosyl-tRNA synthetase |
| chr19_+_47843409 | 0.76 |
ENSMUST00000026050.8
|
Gsto1
|
glutathione S-transferase omega 1 |
| chr8_-_94262335 | 0.75 |
ENSMUST00000212009.2
ENSMUST00000077816.7 |
Ces5a
|
carboxylesterase 5A |
| chr7_-_103814019 | 0.75 |
ENSMUST00000154555.2
ENSMUST00000051137.15 |
Olfm5
|
olfactomedin 5 |
| chr9_+_106354463 | 0.75 |
ENSMUST00000047721.10
|
Rrp9
|
ribosomal RNA processing 9, U3 small nucleolar RNA binding protein |
| chr13_+_46655617 | 0.75 |
ENSMUST00000225824.2
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr2_-_63014622 | 0.75 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr9_-_54467419 | 0.75 |
ENSMUST00000041901.7
|
Cib2
|
calcium and integrin binding family member 2 |
| chr2_+_26800757 | 0.75 |
ENSMUST00000102898.5
|
Rpl7a
|
ribosomal protein L7A |
| chr5_+_16139683 | 0.74 |
ENSMUST00000167946.9
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr12_+_102915102 | 0.74 |
ENSMUST00000101099.12
|
Unc79
|
unc-79 homolog |
| chr14_-_63402374 | 0.74 |
ENSMUST00000238526.2
|
Fdft1
|
farnesyl diphosphate farnesyl transferase 1 |
| chr16_+_41353360 | 0.74 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr2_+_25313240 | 0.73 |
ENSMUST00000134259.8
ENSMUST00000100320.5 |
Fut7
|
fucosyltransferase 7 |
| chr9_-_57528043 | 0.73 |
ENSMUST00000093832.11
|
Lman1l
|
lectin, mannose-binding 1 like |
| chr7_-_126046814 | 0.73 |
ENSMUST00000146973.2
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr10_+_78870557 | 0.73 |
ENSMUST00000082244.3
|
Olfr57
|
olfactory receptor 57 |
| chr4_+_13784749 | 0.73 |
ENSMUST00000098256.4
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr3_+_5815863 | 0.73 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr1_-_23422230 | 0.73 |
ENSMUST00000027343.6
|
Ogfrl1
|
opioid growth factor receptor-like 1 |
| chr4_-_59915035 | 0.73 |
ENSMUST00000030081.2
|
Slc46a2
|
solute carrier family 46, member 2 |
| chr5_+_31212165 | 0.72 |
ENSMUST00000202795.4
ENSMUST00000201182.4 ENSMUST00000200953.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr9_+_108660989 | 0.72 |
ENSMUST00000192307.6
ENSMUST00000193560.6 ENSMUST00000194875.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr11_+_43046476 | 0.72 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr16_-_4340920 | 0.72 |
ENSMUST00000090500.10
ENSMUST00000023161.8 |
Srl
|
sarcalumenin |
| chrX_+_142301666 | 0.72 |
ENSMUST00000134402.8
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr19_+_46316617 | 0.72 |
ENSMUST00000026256.9
ENSMUST00000177667.2 |
Fbxl15
|
F-box and leucine-rich repeat protein 15 |
| chr5_+_129097133 | 0.71 |
ENSMUST00000031383.14
ENSMUST00000111343.2 |
Ran
|
RAN, member RAS oncogene family |
| chr7_-_44785815 | 0.71 |
ENSMUST00000146760.7
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 2.1 | 18.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.2 | 3.6 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.8 | 9.4 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.7 | 2.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.6 | 1.8 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.6 | 2.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.5 | 3.8 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.5 | 1.6 | GO:0061193 | taste bud development(GO:0061193) |
| 0.4 | 0.4 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.4 | 2.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.4 | 1.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 | 2.4 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.4 | 1.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.4 | 1.5 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.4 | 1.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.4 | 1.8 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.4 | 1.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.3 | 1.0 | GO:1904434 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.3 | 5.0 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.3 | 0.3 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.3 | 0.9 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.3 | 1.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.3 | 0.9 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.3 | 1.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.3 | 1.5 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.3 | 0.9 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 0.9 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.3 | 3.7 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.3 | 1.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.3 | 0.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 | 0.3 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.3 | 3.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 2.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.3 | 1.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.3 | 0.8 | GO:0097273 | creatinine homeostasis(GO:0097273) cellular ammonia homeostasis(GO:0097275) cellular creatinine homeostasis(GO:0097276) cellular urea homeostasis(GO:0097277) |
| 0.3 | 1.0 | GO:0002465 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) |
| 0.3 | 1.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.3 | 1.3 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.3 | 5.0 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.2 | 8.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 9.7 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.2 | 4.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 | 0.7 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 2.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 1.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.7 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.2 | 0.7 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.2 | 1.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.2 | 0.7 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 | 0.7 | GO:0060197 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) cloacal septation(GO:0060197) |
| 0.2 | 0.9 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 1.3 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 0.6 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 0.6 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.2 | 1.0 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.2 | 0.6 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.2 | 0.6 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 1.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 0.8 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 0.8 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 1.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.2 | 0.8 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 | 0.8 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.2 | 1.1 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.2 | 0.6 | GO:0032976 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.2 | 1.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 1.3 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.2 | 0.4 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 | 0.4 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.2 | 0.5 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.2 | 1.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 0.7 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.2 | 0.7 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.2 | 0.5 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.2 | 0.5 | GO:0002930 | trabecular meshwork development(GO:0002930) endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 0.5 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.2 | 0.5 | GO:0061723 | glycophagy(GO:0061723) |
| 0.2 | 0.5 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.2 | 0.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.2 | 1.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 1.0 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.2 | 0.3 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.2 | 0.9 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.2 | 0.8 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 1.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.5 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 1.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.4 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.9 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.1 | 1.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.4 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.9 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 2.8 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.6 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.1 | 0.6 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.1 | 0.4 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.8 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.3 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 0.1 | 0.4 | GO:0060032 | floor plate formation(GO:0021508) notochord regression(GO:0060032) |
| 0.1 | 0.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.4 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 0.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.4 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.1 | 0.4 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.1 | 0.8 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.0 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 0.4 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.1 | 1.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.5 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.4 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.6 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.9 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.5 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.7 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.6 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.4 | GO:0021750 | vestibular nucleus development(GO:0021750) |
| 0.1 | 3.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.5 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.6 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.7 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.7 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 1.1 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.1 | 0.5 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.1 | 0.6 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.4 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.3 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 1.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.4 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 0.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 1.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.1 | 0.4 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 1.6 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.3 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 0.5 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.9 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.0 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.9 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.4 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 | 0.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.6 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.3 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.3 | GO:1903632 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.1 | 0.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 1.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.6 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.1 | 0.3 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.8 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.3 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.1 | 0.6 | GO:0043622 | cortical microtubule organization(GO:0043622) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.4 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.6 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 2.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.5 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.3 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 2.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.3 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 1.5 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.3 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 1.0 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.1 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.3 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 | 2.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 0.6 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.3 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.5 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.7 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.9 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.6 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.9 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.2 | GO:0046436 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 1.8 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 | 0.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 1.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.4 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 1.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.4 | GO:0015812 | gamma-aminobutyric acid secretion(GO:0014051) gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.4 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.4 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.1 | 0.4 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.3 | GO:0060031 | positive regulation of transforming growth factor beta2 production(GO:0032915) mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.1 | 0.8 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.2 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) |
| 0.1 | 1.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.5 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 0.2 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 0.4 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.6 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.1 | 0.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.4 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.2 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.6 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.5 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.9 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.3 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.1 | 0.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.4 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.8 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.1 | 0.4 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 1.0 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.5 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.8 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 0.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.6 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.5 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.3 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.6 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.3 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 0.6 | GO:0045078 | regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.2 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 2.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.9 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.2 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.8 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 1.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.6 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.7 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 2.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.3 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 0.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.5 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 0.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.3 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 1.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 1.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 1.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 2.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.3 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.7 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.3 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.2 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 34.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.9 | GO:0060444 | branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.0 | 0.3 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.6 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 1.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.4 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.2 | GO:1903286 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 1.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.4 | GO:1990144 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 | 0.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.7 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 1.0 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.4 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.4 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 1.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.5 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 1.3 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 2.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.7 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 1.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0032365 | intracellular lipid transport(GO:0032365) intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.3 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.4 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.0 | GO:1901977 | negative regulation of cell cycle checkpoint(GO:1901977) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.5 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.2 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.1 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.5 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.2 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 1.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.2 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.8 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.9 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.3 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 1.1 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 0.0 | GO:0035874 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 0.0 | 0.5 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.3 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:0032621 | interleukin-18 production(GO:0032621) negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.2 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.0 | GO:1903999 | regulation of eating behavior(GO:1903998) negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.4 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.4 | GO:0032350 | regulation of hormone metabolic process(GO:0032350) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:0060686 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.3 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.2 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:0071548 | response to dexamethasone(GO:0071548) |
| 0.0 | 0.3 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 19.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 1.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.3 | 2.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 0.9 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 2.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 1.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 1.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 0.7 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 1.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 1.9 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 1.8 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 0.6 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.2 | 3.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.6 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 0.8 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 1.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 0.6 | GO:0097144 | BAX complex(GO:0097144) |
| 0.2 | 0.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 0.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 0.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 0.7 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 3.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 1.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 0.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 2.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 4.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.4 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.1 | 0.6 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.1 | 1.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.4 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.4 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.1 | 0.4 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 1.0 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 3.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 2.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 1.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 6.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 8.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.4 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 1.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 3.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.4 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 0.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.8 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.2 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 1.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 1.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 5.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 5.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.0 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 4.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 4.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.5 | GO:1904949 | ATPase complex(GO:1904949) |
| 0.0 | 0.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 3.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 5.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.3 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 0.8 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 3.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 5.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.0 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 2.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.2 | GO:0098793 | presynapse(GO:0098793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.4 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 2.3 | 11.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 2.1 | 18.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.0 | 5.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.8 | 5.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.7 | 2.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.6 | 3.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 1.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 1.5 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.5 | 2.3 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.5 | 1.8 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.4 | 1.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.4 | 1.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.4 | 2.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 1.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.3 | 1.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.3 | 0.9 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 2.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.3 | 1.1 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.3 | 0.3 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.3 | 1.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 1.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.3 | 0.5 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.3 | 1.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 0.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 1.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 2.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 4.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 8.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 2.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 1.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 0.6 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.2 | 1.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 0.8 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 0.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 0.9 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 0.5 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 0.9 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 0.5 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 1.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 2.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 0.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 0.7 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.2 | 1.5 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 0.5 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.8 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.2 | 0.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 0.5 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.2 | 1.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 1.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 5.5 | GO:1905030 | voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.4 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.1 | 0.4 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.4 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 1.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.5 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.9 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.7 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.4 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 0.4 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 1.4 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.1 | 0.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.8 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.1 | 0.4 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 4.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 1.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 1.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 2.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.4 | GO:0031699 | beta-adrenergic receptor activity(GO:0004939) beta-3 adrenergic receptor binding(GO:0031699) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 15.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.5 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.7 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 1.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 1.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.3 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 0.3 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 1.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.3 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 0.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 2.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 2.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.0 | GO:0070330 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.1 | 1.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 33.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.0 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 1.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 2.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.1 | GO:0030594 | neurotransmitter receptor activity(GO:0030594) |
| 0.0 | 0.2 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.6 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0030060 | malate dehydrogenase activity(GO:0016615) L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.5 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.2 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.4 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 1.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.8 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0052597 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.3 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0043142 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 11.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 4.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 5.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 3.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 5.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 14.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 6.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.9 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 0.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.4 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.1 | 3.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 6.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.8 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.1 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.1 | 0.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 3.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 2.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.5 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.7 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |