Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
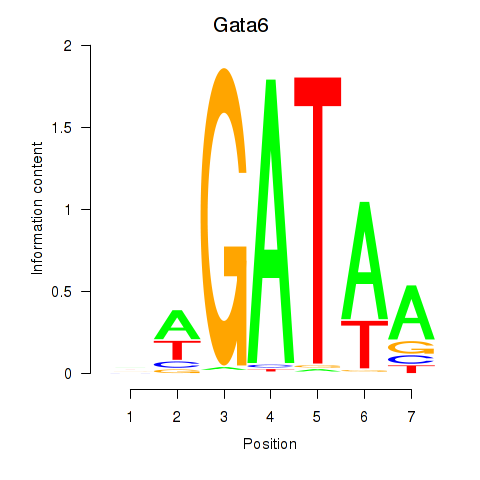
Results for Gata6
Z-value: 0.70
Transcription factors associated with Gata6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata6
|
ENSMUSG00000005836.11 | GATA binding protein 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata6 | mm39_v1_chr18_+_11052458_11052479 | 0.13 | 4.5e-01 | Click! |
Activity profile of Gata6 motif
Sorted Z-values of Gata6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_30639217 | 8.44 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr11_-_102255999 | 8.03 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr4_-_137157824 | 6.38 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr4_-_46404224 | 6.28 |
ENSMUST00000107764.9
|
Hemgn
|
hemogen |
| chr5_+_90920353 | 5.36 |
ENSMUST00000202625.2
|
Pf4
|
platelet factor 4 |
| chr19_+_58658779 | 4.66 |
ENSMUST00000057270.9
|
Pnlip
|
pancreatic lipase |
| chr19_+_58658838 | 4.57 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr10_+_115653152 | 4.50 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr6_+_86055018 | 4.35 |
ENSMUST00000205034.3
ENSMUST00000203724.3 |
Add2
|
adducin 2 (beta) |
| chr15_-_103159892 | 4.25 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr9_-_44253588 | 3.77 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr10_-_62178453 | 3.73 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr6_+_86055048 | 3.50 |
ENSMUST00000032069.8
|
Add2
|
adducin 2 (beta) |
| chr7_-_103502404 | 3.43 |
ENSMUST00000033229.5
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr8_-_86091946 | 2.71 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_-_86091970 | 2.71 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr9_-_44253630 | 2.42 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr13_-_55676334 | 2.30 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr2_-_172212426 | 2.03 |
ENSMUST00000109139.8
ENSMUST00000028997.8 ENSMUST00000109140.10 |
Aurka
|
aurora kinase A |
| chr4_-_137137088 | 2.02 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr19_+_45433899 | 1.89 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr3_-_75177378 | 1.73 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr1_-_144651157 | 1.69 |
ENSMUST00000027603.4
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr2_+_72306503 | 1.57 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr17_+_34823790 | 1.54 |
ENSMUST00000173242.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr7_-_115445352 | 1.53 |
ENSMUST00000206369.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr5_+_76988444 | 1.37 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr11_-_115968373 | 1.21 |
ENSMUST00000174822.8
|
Unc13d
|
unc-13 homolog D |
| chr3_-_20296337 | 1.09 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr3_+_108272205 | 1.06 |
ENSMUST00000090563.7
|
Mybphl
|
myosin binding protein H-like |
| chr11_-_70111796 | 1.01 |
ENSMUST00000060010.3
ENSMUST00000190533.2 |
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr11_+_97917520 | 0.84 |
ENSMUST00000092425.11
|
Rpl19
|
ribosomal protein L19 |
| chr15_+_57849269 | 0.83 |
ENSMUST00000050374.3
|
Fam83a
|
family with sequence similarity 83, member A |
| chr11_-_115967873 | 0.81 |
ENSMUST00000153408.8
|
Unc13d
|
unc-13 homolog D |
| chr11_+_97917746 | 0.81 |
ENSMUST00000017548.7
|
Rpl19
|
ribosomal protein L19 |
| chr16_+_78727829 | 0.72 |
ENSMUST00000114216.2
ENSMUST00000069148.13 ENSMUST00000023568.14 |
Chodl
|
chondrolectin |
| chr9_+_66257747 | 0.72 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr11_-_99383938 | 0.62 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr15_-_38518458 | 0.60 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr13_+_19398273 | 0.59 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chr2_+_110427643 | 0.56 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr4_+_34893772 | 0.54 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr8_+_57774010 | 0.43 |
ENSMUST00000040104.5
|
Hand2
|
heart and neural crest derivatives expressed 2 |
| chr4_-_132649798 | 0.39 |
ENSMUST00000097856.10
ENSMUST00000030696.11 |
Fam76a
|
family with sequence similarity 76, member A |
| chr1_+_12762501 | 0.35 |
ENSMUST00000177608.8
ENSMUST00000180062.8 |
Sulf1
|
sulfatase 1 |
| chr5_-_73349191 | 0.34 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chrX_+_132751729 | 0.34 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr17_+_71511642 | 0.26 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr2_+_109522781 | 0.26 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr3_+_90201388 | 0.25 |
ENSMUST00000199607.5
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr2_+_172212577 | 0.24 |
ENSMUST00000151511.8
|
Cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr1_-_160134873 | 0.18 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr16_+_91444730 | 0.15 |
ENSMUST00000119368.8
ENSMUST00000114037.9 ENSMUST00000114036.9 ENSMUST00000122302.8 |
Son
|
Son DNA binding protein |
| chr10_+_101517348 | 0.15 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr18_+_76192529 | 0.14 |
ENSMUST00000167921.2
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr9_+_64939695 | 0.13 |
ENSMUST00000034960.14
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr5_-_30619246 | 0.13 |
ENSMUST00000114747.9
ENSMUST00000074171.10 |
Otof
|
otoferlin |
| chr7_+_100435548 | 0.12 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A |
| chr16_-_19334384 | 0.12 |
ENSMUST00000054606.2
|
Olfr167
|
olfactory receptor 167 |
| chr2_-_59778560 | 0.11 |
ENSMUST00000153136.2
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chrX_-_111608339 | 0.10 |
ENSMUST00000039887.4
|
Pof1b
|
premature ovarian failure 1B |
| chr1_-_131161312 | 0.09 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr2_-_125701059 | 0.08 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr9_-_95697441 | 0.08 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr11_-_53598078 | 0.06 |
ENSMUST00000124352.2
ENSMUST00000020649.14 |
Rad50
|
RAD50 double strand break repair protein |
| chr13_-_22403990 | 0.02 |
ENSMUST00000057516.2
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr13_-_103042554 | 0.01 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr15_-_73114855 | 0.01 |
ENSMUST00000227686.2
|
Ptk2
|
PTK2 protein tyrosine kinase 2 |
| chr16_+_35861554 | 0.00 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chr2_-_7400690 | 0.00 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 1.1 | 5.4 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.9 | 5.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.7 | 2.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.5 | 3.7 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 7.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.4 | 3.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 8.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 4.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.3 | 2.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.2 | 7.9 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.6 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 1.5 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 1.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.3 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 1.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 4.5 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.4 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 1.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 8.0 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.7 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 3.8 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.4 | 3.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 5.4 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 2.0 | GO:0042585 | chromosome passenger complex(GO:0032133) germinal vesicle(GO:0042585) |
| 0.2 | 2.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 3.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 8.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 6.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.9 | 5.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.9 | 3.4 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.6 | 5.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 3.7 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.5 | 9.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 1.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.4 | 8.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 9.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 2.0 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 7.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 4.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 8.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 2.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 4.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.9 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 9.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 6.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 9.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 8.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 1.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |