Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
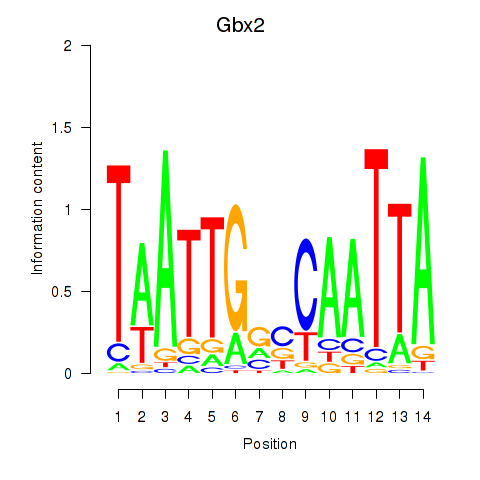
Results for Gbx2
Z-value: 0.58
Transcription factors associated with Gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx2
|
ENSMUSG00000034486.9 | gastrulation brain homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gbx2 | mm39_v1_chr1_-_89858901_89858920 | -0.49 | 2.4e-03 | Click! |
Activity profile of Gbx2 motif
Sorted Z-values of Gbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_38995463 | 2.90 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr19_+_38121248 | 1.48 |
ENSMUST00000025956.13
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr17_-_36343573 | 1.36 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr19_+_38121214 | 1.36 |
ENSMUST00000112329.3
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr7_-_141014477 | 1.27 |
ENSMUST00000106007.10
ENSMUST00000150026.2 ENSMUST00000202840.4 ENSMUST00000133206.9 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr3_+_94280101 | 1.05 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr7_-_141014336 | 1.01 |
ENSMUST00000136354.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr7_-_141014445 | 0.98 |
ENSMUST00000133021.2
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr7_-_141014192 | 0.98 |
ENSMUST00000201127.5
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chrM_-_14061 | 0.96 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr3_-_113371392 | 0.92 |
ENSMUST00000067980.12
|
Amy1
|
amylase 1, salivary |
| chr11_-_21320452 | 0.88 |
ENSMUST00000102875.11
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr5_-_31453206 | 0.87 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chrM_+_14138 | 0.82 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr11_-_46597885 | 0.79 |
ENSMUST00000055102.13
ENSMUST00000125008.2 |
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr9_+_106325860 | 0.78 |
ENSMUST00000185527.3
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr11_+_97206542 | 0.75 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr3_+_57332735 | 0.70 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr2_+_26969384 | 0.64 |
ENSMUST00000091233.7
|
Adamtsl2
|
ADAMTS-like 2 |
| chr9_+_50466127 | 0.59 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr1_-_175453117 | 0.58 |
ENSMUST00000027810.14
|
Fh1
|
fumarate hydratase 1 |
| chr4_-_139695337 | 0.50 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr5_+_113873864 | 0.46 |
ENSMUST00000065698.7
|
Ficd
|
FIC domain containing |
| chr6_+_41118120 | 0.45 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr14_+_69585036 | 0.44 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr11_+_99748741 | 0.41 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr10_-_95337783 | 0.31 |
ENSMUST00000075829.3
ENSMUST00000217777.2 ENSMUST00000218893.2 |
Mrpl42
|
mitochondrial ribosomal protein L42 |
| chr2_-_157408239 | 0.25 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr10_-_116732813 | 0.22 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr2_-_17465410 | 0.21 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr1_-_24044688 | 0.20 |
ENSMUST00000027338.4
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr1_-_128030148 | 0.19 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr4_-_129155185 | 0.18 |
ENSMUST00000145261.8
|
C77080
|
expressed sequence C77080 |
| chr9_+_21634779 | 0.17 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr16_-_26190578 | 0.16 |
ENSMUST00000023154.3
|
Cldn1
|
claudin 1 |
| chr5_+_7354130 | 0.16 |
ENSMUST00000160634.2
ENSMUST00000159546.2 |
Tex47
|
testis expressed 47 |
| chr5_+_117919082 | 0.14 |
ENSMUST00000138579.3
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr1_-_74932266 | 0.14 |
ENSMUST00000006721.3
|
Cryba2
|
crystallin, beta A2 |
| chr7_-_78432774 | 0.12 |
ENSMUST00000032841.7
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr5_-_5315968 | 0.12 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr17_-_37974666 | 0.12 |
ENSMUST00000215414.2
ENSMUST00000213638.3 |
Olfr117
|
olfactory receptor 117 |
| chr14_-_66315144 | 0.11 |
ENSMUST00000022618.6
|
Adam2
|
a disintegrin and metallopeptidase domain 2 |
| chr19_+_17114108 | 0.10 |
ENSMUST00000223920.2
ENSMUST00000225351.2 |
Prune2
|
prune homolog 2 |
| chr3_-_89905547 | 0.09 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr17_-_71153283 | 0.09 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr11_+_59197746 | 0.09 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr16_+_44215136 | 0.09 |
ENSMUST00000099742.9
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr19_-_39729431 | 0.09 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr10_+_116013256 | 0.09 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr10_+_127919142 | 0.08 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr6_-_130170075 | 0.08 |
ENSMUST00000112032.8
ENSMUST00000071554.3 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr1_+_92533504 | 0.08 |
ENSMUST00000217316.2
ENSMUST00000216553.2 |
Olfr1410
|
olfactory receptor 1410 |
| chr11_-_99742434 | 0.08 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr3_-_10396418 | 0.08 |
ENSMUST00000191670.6
ENSMUST00000065938.15 ENSMUST00000118410.8 |
Impa1
|
inositol (myo)-1(or 4)-monophosphatase 1 |
| chr9_+_108708939 | 0.08 |
ENSMUST00000192235.2
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr2_-_85632888 | 0.07 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr2_+_70392491 | 0.07 |
ENSMUST00000148210.8
|
Gad1
|
glutamate decarboxylase 1 |
| chr18_-_78640066 | 0.07 |
ENSMUST00000235389.2
ENSMUST00000237674.2 |
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr11_-_58521327 | 0.07 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr1_-_4855894 | 0.06 |
ENSMUST00000130201.8
ENSMUST00000156816.7 ENSMUST00000146665.3 |
Mrpl15
|
mitochondrial ribosomal protein L15 |
| chr17_-_33028814 | 0.06 |
ENSMUST00000234594.2
|
Zfp811
|
zinc finger protein 811 |
| chr10_+_116013122 | 0.05 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_161807205 | 0.05 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr17_+_38374812 | 0.05 |
ENSMUST00000215726.2
ENSMUST00000217390.2 |
Olfr130
|
olfactory receptor 130 |
| chr4_+_52596266 | 0.05 |
ENSMUST00000029995.6
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr14_+_45225641 | 0.04 |
ENSMUST00000046891.6
|
Ptger2
|
prostaglandin E receptor 2 (subtype EP2) |
| chr2_+_70392351 | 0.04 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chr2_-_111324108 | 0.03 |
ENSMUST00000208881.2
ENSMUST00000208695.2 ENSMUST00000217611.2 |
Olfr1290
|
olfactory receptor 1290 |
| chr6_+_114435480 | 0.03 |
ENSMUST00000160780.2
|
Hrh1
|
histamine receptor H1 |
| chr14_-_48904958 | 0.03 |
ENSMUST00000144465.8
ENSMUST00000133479.8 ENSMUST00000119070.8 ENSMUST00000226501.2 |
Otx2
|
orthodenticle homeobox 2 |
| chr8_+_22329942 | 0.03 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chrX_-_42256694 | 0.03 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr8_-_41494890 | 0.02 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr7_+_28510310 | 0.01 |
ENSMUST00000038572.15
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr3_-_89905927 | 0.01 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr1_+_179936757 | 0.01 |
ENSMUST00000143176.8
ENSMUST00000135056.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_129077867 | 0.01 |
ENSMUST00000032258.8
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr11_-_73707875 | 0.01 |
ENSMUST00000118463.4
ENSMUST00000144724.3 |
Olfr392
|
olfactory receptor 392 |
| chr14_-_50390356 | 0.01 |
ENSMUST00000215451.2
ENSMUST00000213163.2 ENSMUST00000215327.2 |
Olfr729
|
olfactory receptor 729 |
| chr2_-_73410632 | 0.01 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr17_+_85264134 | 0.01 |
ENSMUST00000112305.10
|
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr2_+_113116075 | 0.00 |
ENSMUST00000040856.3
|
Tmco5b
|
transmembrane and coiled-coil domains 5B |
| chr8_-_22396428 | 0.00 |
ENSMUST00000051965.5
|
Defb11
|
defensin beta 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:2000566 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.3 | 0.9 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 | 2.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 0.6 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.2 | 2.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.6 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 4.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.8 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 1.1 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.6 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.8 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 1.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.8 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.6 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 1.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.9 | GO:0070469 | respiratory chain(GO:0070469) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.3 | 4.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 1.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 0.8 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.1 | 0.9 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.9 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.4 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |