Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
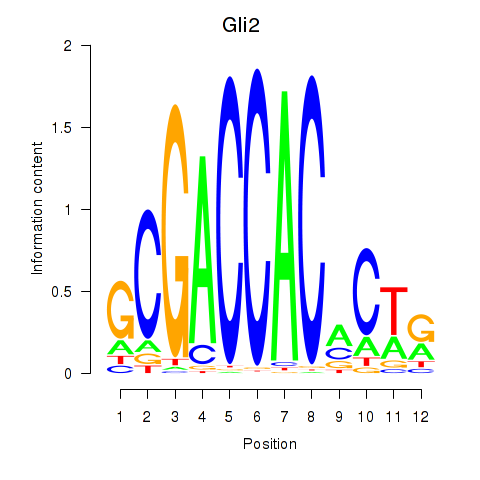
Results for Gli2
Z-value: 0.87
Transcription factors associated with Gli2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli2
|
ENSMUSG00000048402.15 | GLI-Kruppel family member GLI2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli2 | mm39_v1_chr1_-_118981341_118981368 | -0.77 | 3.8e-08 | Click! |
Activity profile of Gli2 motif
Sorted Z-values of Gli2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_51955054 | 5.15 |
ENSMUST00000018561.14
ENSMUST00000114537.9 |
Myo1b
|
myosin IB |
| chr3_+_97536120 | 4.98 |
ENSMUST00000107050.8
ENSMUST00000029729.15 ENSMUST00000107049.2 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr4_-_154983533 | 4.85 |
ENSMUST00000030935.10
ENSMUST00000132281.2 |
Prxl2b
|
peroxiredoxin like 2B |
| chr5_-_53370761 | 4.81 |
ENSMUST00000031090.8
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr1_-_51955126 | 4.80 |
ENSMUST00000046390.14
|
Myo1b
|
myosin IB |
| chr9_+_107454114 | 3.80 |
ENSMUST00000112387.9
ENSMUST00000123005.8 ENSMUST00000010195.14 ENSMUST00000144392.2 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr1_-_180027151 | 3.59 |
ENSMUST00000161743.3
|
Coq8a
|
coenzyme Q8A |
| chr8_+_127790772 | 3.40 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr9_+_100525807 | 3.22 |
ENSMUST00000133388.2
|
Stag1
|
stromal antigen 1 |
| chr4_-_120955387 | 2.84 |
ENSMUST00000058754.9
|
Zmpste24
|
zinc metallopeptidase, STE24 |
| chr9_+_100525637 | 2.62 |
ENSMUST00000041418.13
|
Stag1
|
stromal antigen 1 |
| chr2_-_157408239 | 2.49 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr17_-_24428351 | 2.31 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr2_-_32271833 | 2.25 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr2_-_121786573 | 2.19 |
ENSMUST00000104936.4
|
Mageb3
|
MAGE family member B3 |
| chr12_-_70274433 | 2.19 |
ENSMUST00000071250.13
|
Pygl
|
liver glycogen phosphorylase |
| chr2_+_69727599 | 2.18 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr4_-_46536088 | 2.09 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr9_+_100525501 | 2.09 |
ENSMUST00000146312.8
ENSMUST00000129269.8 |
Stag1
|
stromal antigen 1 |
| chr9_+_108924457 | 2.07 |
ENSMUST00000072093.13
|
Plxnb1
|
plexin B1 |
| chr1_-_93406091 | 1.99 |
ENSMUST00000188165.2
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr11_+_118913788 | 1.99 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr6_+_137731526 | 1.90 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr8_+_91635192 | 1.87 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chrX_-_9529189 | 1.79 |
ENSMUST00000033519.3
|
Dynlt3
|
dynein light chain Tctex-type 3 |
| chr8_+_77628916 | 1.78 |
ENSMUST00000109912.8
ENSMUST00000128862.2 ENSMUST00000109911.8 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr7_-_81104423 | 1.75 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr16_+_4825216 | 1.74 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr6_+_113448388 | 1.74 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr6_+_137731599 | 1.74 |
ENSMUST00000204356.2
|
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr19_-_38113249 | 1.71 |
ENSMUST00000112335.4
|
Rbp4
|
retinol binding protein 4, plasma |
| chr1_-_175453117 | 1.64 |
ENSMUST00000027810.14
|
Fh1
|
fumarate hydratase 1 |
| chr2_+_160573604 | 1.62 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr4_+_41760454 | 1.60 |
ENSMUST00000108040.8
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr10_-_127099183 | 1.56 |
ENSMUST00000099172.5
|
Kif5a
|
kinesin family member 5A |
| chr7_+_28240262 | 1.50 |
ENSMUST00000119180.4
|
Sycn
|
syncollin |
| chrX_-_73067514 | 1.50 |
ENSMUST00000033769.15
ENSMUST00000114352.8 ENSMUST00000068286.12 ENSMUST00000114360.10 ENSMUST00000114354.10 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr11_+_72686990 | 1.48 |
ENSMUST00000069395.7
ENSMUST00000172220.8 |
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr10_+_83379805 | 1.45 |
ENSMUST00000038388.7
|
Washc4
|
WASH complex subunit 4 |
| chr2_-_37593287 | 1.44 |
ENSMUST00000072186.12
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr12_-_69771604 | 1.43 |
ENSMUST00000021370.10
|
L2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr7_-_65020955 | 1.43 |
ENSMUST00000102592.10
|
Tjp1
|
tight junction protein 1 |
| chr10_-_127098932 | 1.41 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr13_-_54759086 | 1.40 |
ENSMUST00000049575.8
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr11_+_70735751 | 1.39 |
ENSMUST00000177731.8
ENSMUST00000108533.10 ENSMUST00000081362.13 ENSMUST00000178245.2 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr19_+_6097111 | 1.38 |
ENSMUST00000025723.9
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr1_-_93406515 | 1.36 |
ENSMUST00000170883.8
ENSMUST00000189025.7 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr19_-_38113056 | 1.33 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr19_+_6097083 | 1.32 |
ENSMUST00000134667.8
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr2_+_121786892 | 1.32 |
ENSMUST00000110578.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr8_+_120173458 | 1.29 |
ENSMUST00000098363.10
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr13_-_54759145 | 1.27 |
ENSMUST00000091609.11
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr2_-_121211410 | 1.23 |
ENSMUST00000038389.15
|
Strc
|
stereocilin |
| chrX_+_160500623 | 1.22 |
ENSMUST00000061514.8
|
Rai2
|
retinoic acid induced 2 |
| chr15_+_41310815 | 1.21 |
ENSMUST00000090096.11
ENSMUST00000110297.10 |
Oxr1
|
oxidation resistance 1 |
| chr9_+_44966464 | 1.20 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr12_-_32000209 | 1.19 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr8_+_127790626 | 1.18 |
ENSMUST00000162309.8
|
Pard3
|
par-3 family cell polarity regulator |
| chr7_+_81220987 | 1.14 |
ENSMUST00000165460.2
|
Whamm
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr4_-_140376047 | 1.13 |
ENSMUST00000105799.8
ENSMUST00000039204.10 ENSMUST00000097820.9 |
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr4_+_155916087 | 1.10 |
ENSMUST00000105592.8
ENSMUST00000105591.2 |
Aurkaip1
|
aurora kinase A interacting protein 1 |
| chr11_-_121279062 | 1.08 |
ENSMUST00000106107.3
|
Rab40b
|
Rab40B, member RAS oncogene family |
| chr3_+_106943472 | 1.08 |
ENSMUST00000052718.5
|
Kcna3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr9_+_100956145 | 1.07 |
ENSMUST00000189616.2
|
Msl2
|
MSL complex subunit 2 |
| chr17_+_24851647 | 1.05 |
ENSMUST00000047611.4
|
Nthl1
|
nth (endonuclease III)-like 1 (E.coli) |
| chr1_+_167177545 | 1.05 |
ENSMUST00000028004.11
|
Aldh9a1
|
aldehyde dehydrogenase 9, subfamily A1 |
| chr5_-_139805661 | 1.04 |
ENSMUST00000147328.2
|
Tmem184a
|
transmembrane protein 184a |
| chr12_-_32000169 | 1.03 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr7_-_144292257 | 0.94 |
ENSMUST00000121758.8
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr14_-_65187287 | 0.92 |
ENSMUST00000067843.10
ENSMUST00000176489.8 ENSMUST00000175905.8 ENSMUST00000022544.14 ENSMUST00000175744.8 ENSMUST00000176128.8 |
Hmbox1
|
homeobox containing 1 |
| chr11_+_70735572 | 0.90 |
ENSMUST00000076270.13
ENSMUST00000179114.8 ENSMUST00000100928.11 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr17_+_28910393 | 0.89 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr11_+_29476423 | 0.89 |
ENSMUST00000136351.8
ENSMUST00000020749.13 ENSMUST00000144321.8 ENSMUST00000093239.11 |
Mtif2
|
mitochondrial translational initiation factor 2 |
| chr8_-_27664651 | 0.89 |
ENSMUST00000054212.7
ENSMUST00000033878.14 ENSMUST00000209377.2 |
Rab11fip1
|
RAB11 family interacting protein 1 (class I) |
| chr1_-_172418058 | 0.87 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8 |
| chr1_-_36484328 | 0.87 |
ENSMUST00000125304.8
ENSMUST00000115011.8 |
Lman2l
|
lectin, mannose-binding 2-like |
| chrX_-_140508177 | 0.84 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr8_+_85786684 | 0.84 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr18_+_61688329 | 0.83 |
ENSMUST00000165123.8
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr7_-_65020655 | 0.82 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr11_+_29476385 | 0.82 |
ENSMUST00000133452.8
|
Mtif2
|
mitochondrial translational initiation factor 2 |
| chr16_+_19579651 | 0.82 |
ENSMUST00000119468.2
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr7_+_101034604 | 0.81 |
ENSMUST00000130016.8
ENSMUST00000134143.2 |
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr7_+_40547608 | 0.80 |
ENSMUST00000044705.12
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr8_-_13939964 | 0.80 |
ENSMUST00000209371.2
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr11_+_72687080 | 0.79 |
ENSMUST00000207107.2
|
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr13_-_48779047 | 0.74 |
ENSMUST00000222028.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr7_-_144292288 | 0.74 |
ENSMUST00000238848.2
ENSMUST00000118556.9 ENSMUST00000033393.15 |
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr5_-_92231517 | 0.72 |
ENSMUST00000202258.4
ENSMUST00000113127.7 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr4_+_43401232 | 0.72 |
ENSMUST00000125399.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr17_-_56916771 | 0.71 |
ENSMUST00000052832.6
|
Micos13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr18_-_35348049 | 0.71 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr11_+_106256298 | 0.70 |
ENSMUST00000009354.10
|
Prr29
|
proline rich 29 |
| chr7_-_132724344 | 0.67 |
ENSMUST00000167218.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chrX_+_160500051 | 0.64 |
ENSMUST00000112338.2
|
Rai2
|
retinoic acid induced 2 |
| chr3_-_59102517 | 0.61 |
ENSMUST00000200095.2
|
Gpr87
|
G protein-coupled receptor 87 |
| chr8_-_13940234 | 0.61 |
ENSMUST00000033839.9
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr17_+_45874800 | 0.59 |
ENSMUST00000224905.2
ENSMUST00000226086.2 ENSMUST00000041353.7 |
Slc35b2
|
solute carrier family 35, member B2 |
| chr2_+_180141686 | 0.59 |
ENSMUST00000029084.9
|
Ntsr1
|
neurotensin receptor 1 |
| chr11_+_68582897 | 0.59 |
ENSMUST00000108673.8
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr10_-_34003933 | 0.58 |
ENSMUST00000062784.8
|
Calhm6
|
calcium homeostasis modulator family member 6 |
| chr7_-_132725041 | 0.57 |
ENSMUST00000171022.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr11_-_94595325 | 0.57 |
ENSMUST00000069852.2
|
Gm11541
|
predicted gene 11541 |
| chr13_-_48779072 | 0.54 |
ENSMUST00000035824.11
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr7_-_57159119 | 0.53 |
ENSMUST00000206382.2
|
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr11_+_73220553 | 0.52 |
ENSMUST00000092926.11
ENSMUST00000117445.2 |
Spata22
Gm49340
|
spermatogenesis associated 22 predicted gene, 49340 |
| chrX_+_7439839 | 0.52 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chrX_-_166165743 | 0.51 |
ENSMUST00000026839.5
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr16_+_8647959 | 0.50 |
ENSMUST00000023150.7
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chr8_+_10056631 | 0.49 |
ENSMUST00000207792.3
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr17_-_25716189 | 0.48 |
ENSMUST00000165183.10
ENSMUST00000051864.5 |
Sstr5
|
somatostatin receptor 5 |
| chr12_-_32000534 | 0.48 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr18_+_61688378 | 0.48 |
ENSMUST00000165721.8
ENSMUST00000115246.9 ENSMUST00000166990.8 ENSMUST00000163205.8 ENSMUST00000170862.8 |
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr3_-_85909798 | 0.48 |
ENSMUST00000061343.4
|
Prss48
|
protease, serine 48 |
| chr7_-_132725137 | 0.48 |
ENSMUST00000170459.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr11_+_70506716 | 0.43 |
ENSMUST00000144960.2
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr11_-_100741550 | 0.42 |
ENSMUST00000004143.3
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr18_+_35347983 | 0.42 |
ENSMUST00000235449.2
ENSMUST00000235269.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr2_+_164328375 | 0.41 |
ENSMUST00000069385.15
ENSMUST00000143690.8 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_+_70453666 | 0.41 |
ENSMUST00000072237.13
ENSMUST00000072873.14 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr15_-_78602313 | 0.40 |
ENSMUST00000229441.2
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr11_+_115921129 | 0.40 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr9_+_62249730 | 0.40 |
ENSMUST00000156461.2
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr2_+_164327988 | 0.39 |
ENSMUST00000109350.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr12_+_38830081 | 0.36 |
ENSMUST00000095767.11
|
Etv1
|
ets variant 1 |
| chr5_+_8943677 | 0.36 |
ENSMUST00000003717.13
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr8_+_10056654 | 0.35 |
ENSMUST00000033892.9
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr7_+_12615091 | 0.35 |
ENSMUST00000144578.2
|
Zfp128
|
zinc finger protein 128 |
| chr5_-_24732200 | 0.35 |
ENSMUST00000088311.6
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr16_-_3895642 | 0.34 |
ENSMUST00000006137.9
|
Trap1
|
TNF receptor-associated protein 1 |
| chr16_-_31133622 | 0.34 |
ENSMUST00000115230.2
ENSMUST00000130560.8 |
Apod
|
apolipoprotein D |
| chr6_-_124441731 | 0.34 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr1_-_119924920 | 0.34 |
ENSMUST00000174370.8
ENSMUST00000174458.2 |
Cfap221
|
cilia and flagella associated protein 221 |
| chr8_+_71207326 | 0.33 |
ENSMUST00000110093.9
ENSMUST00000143118.3 ENSMUST00000034301.12 ENSMUST00000110090.8 |
Rab3a
|
RAB3A, member RAS oncogene family |
| chr11_+_70453724 | 0.32 |
ENSMUST00000102559.11
|
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr8_+_27937691 | 0.32 |
ENSMUST00000081321.5
|
Poteg
|
POTE ankyrin domain family, member G |
| chr7_+_44813363 | 0.32 |
ENSMUST00000085374.7
ENSMUST00000209634.2 |
Slc17a7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 |
| chr11_-_68762664 | 0.32 |
ENSMUST00000101017.9
|
Ndel1
|
nudE neurodevelopment protein 1 like 1 |
| chr2_+_121786444 | 0.31 |
ENSMUST00000036647.13
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr4_-_126150066 | 0.30 |
ENSMUST00000122129.8
ENSMUST00000061143.15 ENSMUST00000106132.3 |
Map7d1
|
MAP7 domain containing 1 |
| chr7_+_130294262 | 0.29 |
ENSMUST00000033141.7
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chrX_-_95000496 | 0.29 |
ENSMUST00000079987.13
ENSMUST00000113864.3 |
Las1l
|
LAS1-like (S. cerevisiae) |
| chr10_-_128727542 | 0.29 |
ENSMUST00000026408.7
|
Gdf11
|
growth differentiation factor 11 |
| chr7_+_140908033 | 0.28 |
ENSMUST00000133012.8
ENSMUST00000026578.14 |
Tmem80
|
transmembrane protein 80 |
| chr11_+_117198958 | 0.26 |
ENSMUST00000153668.8
|
Septin9
|
septin 9 |
| chr2_-_104542467 | 0.25 |
ENSMUST00000111118.8
ENSMUST00000028597.4 |
Tcp11l1
|
t-complex 11 like 1 |
| chr5_-_71253107 | 0.25 |
ENSMUST00000197284.5
|
Gabra2
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
| chr5_+_147206769 | 0.24 |
ENSMUST00000085591.7
|
Pdx1
|
pancreatic and duodenal homeobox 1 |
| chr17_-_84495364 | 0.24 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr11_+_70453806 | 0.24 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr6_-_38614155 | 0.24 |
ENSMUST00000096030.7
ENSMUST00000201345.2 |
Klrg2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr17_-_52139693 | 0.23 |
ENSMUST00000144331.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr2_+_74566740 | 0.23 |
ENSMUST00000111982.8
|
Hoxd3
|
homeobox D3 |
| chr14_-_55354392 | 0.21 |
ENSMUST00000022819.13
|
Jph4
|
junctophilin 4 |
| chr5_+_73563418 | 0.20 |
ENSMUST00000031040.13
ENSMUST00000065543.8 |
Cwh43
|
cell wall biogenesis 43 C-terminal homolog |
| chr4_-_135699205 | 0.19 |
ENSMUST00000105852.8
|
Lypla2
|
lysophospholipase 2 |
| chr4_+_63133639 | 0.18 |
ENSMUST00000036300.13
|
Col27a1
|
collagen, type XXVII, alpha 1 |
| chr7_-_132725075 | 0.18 |
ENSMUST00000163601.8
ENSMUST00000033269.15 ENSMUST00000124096.8 |
Ctbp2
Fgfr2
|
C-terminal binding protein 2 fibroblast growth factor receptor 2 |
| chrX_+_99773784 | 0.18 |
ENSMUST00000113744.2
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr1_-_72576089 | 0.18 |
ENSMUST00000047786.6
|
Marchf4
|
membrane associated ring-CH-type finger 4 |
| chr4_+_116953218 | 0.15 |
ENSMUST00000030443.12
|
Ptch2
|
patched 2 |
| chr18_+_38383297 | 0.12 |
ENSMUST00000025314.7
ENSMUST00000236078.2 |
Dele1
|
DAP3 binding cell death enhancer 1 |
| chr9_+_107805647 | 0.12 |
ENSMUST00000085073.2
|
Actl11
|
actin-like 11 |
| chr2_-_120439764 | 0.11 |
ENSMUST00000102496.8
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr11_-_41891359 | 0.10 |
ENSMUST00000070735.10
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr2_-_120439981 | 0.09 |
ENSMUST00000133612.2
ENSMUST00000102498.8 ENSMUST00000102499.8 |
Lrrc57
|
leucine rich repeat containing 57 |
| chr9_+_24009292 | 0.09 |
ENSMUST00000133787.8
ENSMUST00000059650.11 |
Npsr1
|
neuropeptide S receptor 1 |
| chr16_+_3648742 | 0.08 |
ENSMUST00000214238.2
ENSMUST00000214590.2 |
Olfr15
|
olfactory receptor 15 |
| chr2_-_120439826 | 0.08 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr12_+_38830283 | 0.07 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chr1_+_55127110 | 0.06 |
ENSMUST00000075242.7
|
Hspe1
|
heat shock protein 1 (chaperonin 10) |
| chr9_+_20943350 | 0.05 |
ENSMUST00000019616.6
|
Icam5
|
intercellular adhesion molecule 5, telencephalin |
| chrX_+_99773523 | 0.05 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr4_+_155915729 | 0.04 |
ENSMUST00000139651.8
ENSMUST00000084097.12 |
Aurkaip1
|
aurora kinase A interacting protein 1 |
| chr1_+_97697881 | 0.04 |
ENSMUST00000112844.10
ENSMUST00000112842.8 ENSMUST00000027571.13 |
Gin1
|
gypsy retrotransposon integrase 1 |
| chr4_+_134879807 | 0.03 |
ENSMUST00000119564.2
|
Runx3
|
runt related transcription factor 3 |
| chr9_-_69668204 | 0.03 |
ENSMUST00000071281.5
|
Foxb1
|
forkhead box B1 |
| chr4_+_40472180 | 0.02 |
ENSMUST00000049655.3
|
Tmem215
|
transmembrane protein 215 |
| chr2_+_71042050 | 0.01 |
ENSMUST00000112142.8
ENSMUST00000112139.8 ENSMUST00000112140.8 ENSMUST00000112138.8 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr11_-_118800314 | 0.00 |
ENSMUST00000117731.8
ENSMUST00000106278.9 ENSMUST00000120061.8 ENSMUST00000017576.11 |
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr17_-_24851549 | 0.00 |
ENSMUST00000227607.2
ENSMUST00000227509.2 ENSMUST00000227745.2 |
Tsc2
|
TSC complex subunit 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.9 | 2.8 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.7 | 2.7 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.7 | 4.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.6 | 3.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.6 | 3.6 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.6 | 3.0 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.5 | 1.6 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.5 | 2.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.5 | 2.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 1.8 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.4 | 1.3 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.4 | 1.1 | GO:0006296 | base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.3 | 0.9 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 0.2 | 1.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 2.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 1.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 1.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.2 | 0.5 | GO:0002851 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.2 | 1.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 3.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 2.7 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 2.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.8 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.4 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.1 | 0.6 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 0.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 1.6 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.3 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.1 | 0.3 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.9 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 3.0 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 1.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.5 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 1.0 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 1.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 10.0 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 1.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 1.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.6 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.6 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 2.5 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.1 | 1.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.4 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.1 | 1.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 1.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 1.3 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.7 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 2.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 1.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.8 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.3 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.0 | GO:0021855 | hypothalamus cell migration(GO:0021855) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 1.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 2.7 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 3.4 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 7.9 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.5 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 10.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 2.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 1.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 1.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 2.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 3.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 2.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.6 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 1.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 3.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 3.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 3.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 2.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.9 | 3.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.6 | 1.8 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.5 | 2.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.5 | 5.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.6 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.3 | 8.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.3 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.3 | 1.7 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.3 | 0.8 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 1.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.2 | 0.6 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 0.6 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.2 | 1.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 2.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 2.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 3.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 2.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.6 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.9 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 1.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.9 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 5.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 3.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 1.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 3.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.0 | 1.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 5.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 3.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 1.3 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.2 | 4.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 7.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 2.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.4 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |