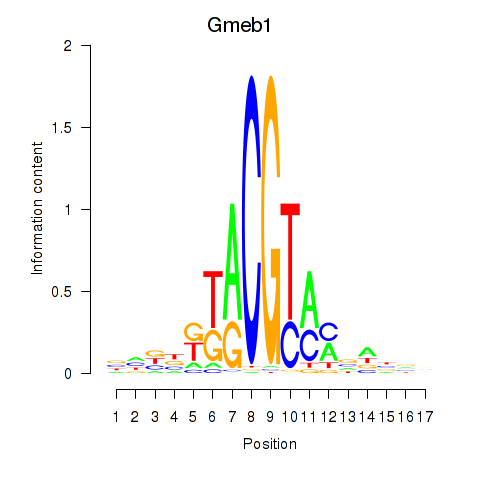
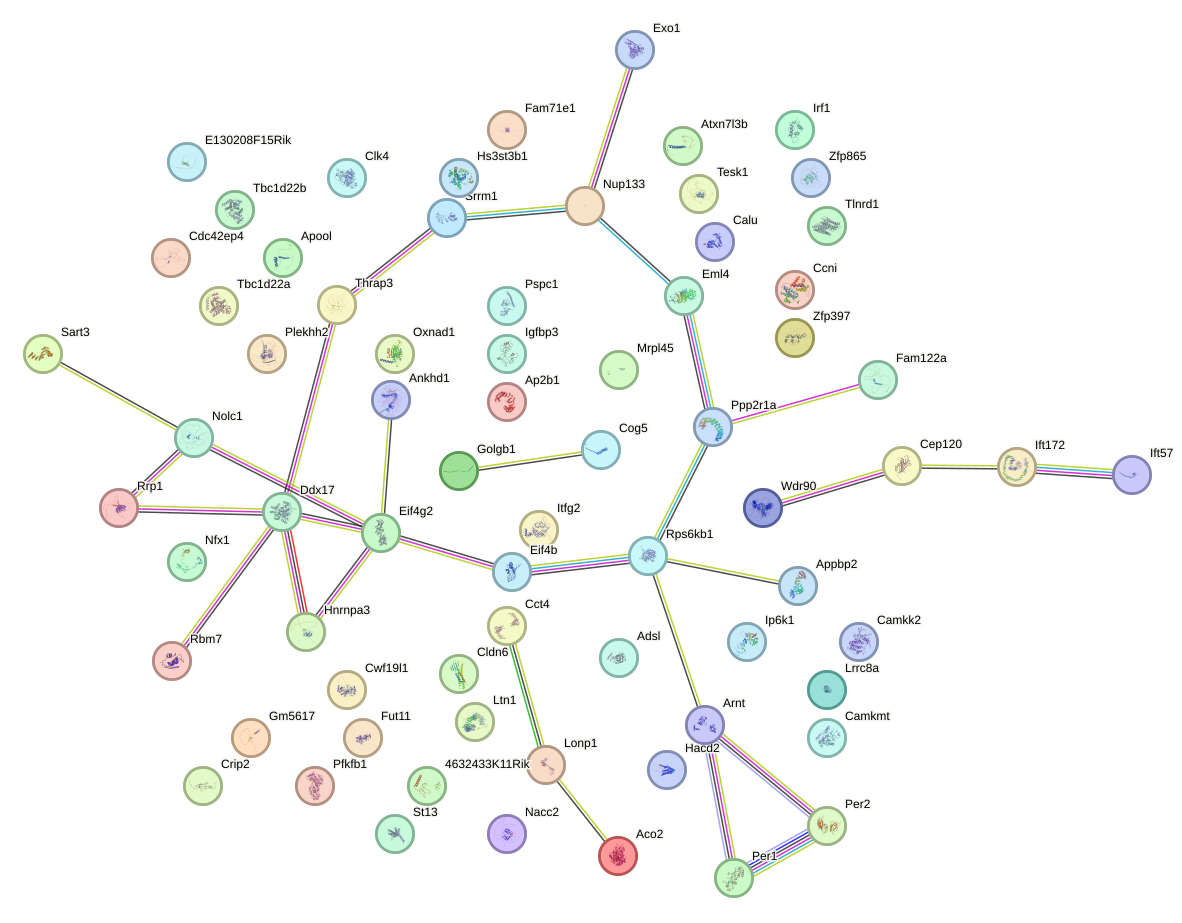
|
chr11_-_7163897
Show fit
|
1.37 |
ENSMUST00000020702.11
ENSMUST00000135887.3
|
Igfbp3
|
insulin-like growth factor binding protein 3
|
|
chr11_+_83193495
Show fit
|
1.11 |
ENSMUST00000176430.8
ENSMUST00000065692.14
ENSMUST00000142680.2
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr1_-_91386976
Show fit
|
0.92 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2
|
|
chr15_-_79430742
Show fit
|
0.91 |
ENSMUST00000231053.2
ENSMUST00000229431.2
|
Ddx17
|
DEAD box helicase 17
|
|
chr4_-_126096376
Show fit
|
0.90 |
ENSMUST00000106142.8
ENSMUST00000169403.8
ENSMUST00000130334.2
|
Thrap3
|
thyroid hormone receptor associated protein 3
|
|
chr8_-_124675939
Show fit
|
0.84 |
ENSMUST00000044795.8
|
Nup133
|
nucleoporin 133
|
|
chr4_-_135080437
Show fit
|
0.83 |
ENSMUST00000030613.11
ENSMUST00000131373.8
|
Srrm1
|
serine/arginine repetitive matrix 1
|
|
chr15_+_81284333
Show fit
|
0.79 |
ENSMUST00000163754.9
ENSMUST00000041609.11
|
Xpnpep3
|
X-prolyl aminopeptidase 3, mitochondrial
|
|
chr2_-_156848923
Show fit
|
0.78 |
ENSMUST00000146413.8
ENSMUST00000103129.9
ENSMUST00000103130.8
|
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component
|
|
chr16_-_87229367
Show fit
|
0.74 |
ENSMUST00000232095.2
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1
|
|
chr11_+_68986043
Show fit
|
0.69 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1
|
|
chr5_-_93354287
Show fit
|
0.63 |
ENSMUST00000144514.3
|
Ccni
|
cyclin I
|
|
chr10_-_112764879
Show fit
|
0.61 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B
|
|
chr3_+_95341698
Show fit
|
0.58 |
ENSMUST00000102749.11
ENSMUST00000090804.12
ENSMUST00000107161.8
ENSMUST00000107160.8
ENSMUST00000015666.17
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr7_+_46495521
Show fit
|
0.57 |
ENSMUST00000133062.2
|
Ldha
|
lactate dehydrogenase A
|
|
chr15_+_81756671
Show fit
|
0.55 |
ENSMUST00000135198.2
ENSMUST00000157003.8
ENSMUST00000229068.2
|
Aco2
|
aconitase 2, mitochondrial
|
|
chr7_-_110682204
Show fit
|
0.54 |
ENSMUST00000161051.8
ENSMUST00000160132.8
ENSMUST00000162415.9
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2
|
|
chr12_+_31704853
Show fit
|
0.52 |
ENSMUST00000036862.5
|
Cog5
|
component of oligomeric golgi complex 5
|
|
chr17_+_83658354
Show fit
|
0.48 |
ENSMUST00000096766.12
ENSMUST00000049503.10
ENSMUST00000112363.10
ENSMUST00000234460.2
|
Eml4
|
echinoderm microtubule associated protein like 4
|
|
chr4_+_40970906
Show fit
|
0.47 |
ENSMUST00000091614.7
ENSMUST00000030133.15
ENSMUST00000098143.11
|
Nfx1
|
nuclear transcription factor, X-box binding 1
|
|
chr14_-_57015748
Show fit
|
0.47 |
ENSMUST00000022507.13
ENSMUST00000163924.2
|
Pspc1
|
paraspeckle protein 1
|
|
chr15_-_81283795
Show fit
|
0.46 |
ENSMUST00000023039.15
|
St13
|
suppression of tumorigenicity 13
|
|
chr18_+_36693024
Show fit
|
0.44 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1
|
|
chr5_-_31448370
Show fit
|
0.44 |
ENSMUST00000041565.11
ENSMUST00000201809.2
|
Ift172
|
intraflagellar transport 172
|
|
chr11_+_53661251
Show fit
|
0.43 |
ENSMUST00000138913.8
ENSMUST00000123376.8
ENSMUST00000019043.13
ENSMUST00000133291.3
|
Irf1
|
interferon regulatory factor 1
|
|
chr9_+_107879700
Show fit
|
0.43 |
ENSMUST00000035214.11
ENSMUST00000176854.7
ENSMUST00000175874.2
|
Ip6k1
|
inositol hexaphosphate kinase 1
|
|
chr7_+_44146029
Show fit
|
0.39 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1
|
|
chr18_+_24087725
Show fit
|
0.39 |
ENSMUST00000225682.2
ENSMUST00000060762.6
|
Zfp397
|
zinc finger protein 397
|
|
chr14_+_20745024
Show fit
|
0.36 |
ENSMUST00000048016.3
|
Fut11
|
fucosyltransferase 11
|
|
chr4_+_43441939
Show fit
|
0.36 |
ENSMUST00000060864.13
|
Tesk1
|
testis specific protein kinase 1
|
|
chr17_+_23898223
Show fit
|
0.36 |
ENSMUST00000024699.4
ENSMUST00000232719.2
|
Cldn6
|
claudin 6
|
|
chr2_+_26899935
Show fit
|
0.36 |
ENSMUST00000114005.9
ENSMUST00000114004.8
ENSMUST00000114006.8
ENSMUST00000114007.8
ENSMUST00000133807.2
|
Cacfd1
|
calcium channel flower domain containing 1
|
|
chr2_+_75489596
Show fit
|
0.35 |
ENSMUST00000111964.8
ENSMUST00000111962.8
ENSMUST00000111961.8
ENSMUST00000164947.9
ENSMUST00000090792.11
|
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3
|
|
chr15_+_101982208
Show fit
|
0.35 |
ENSMUST00000169681.3
ENSMUST00000229400.2
|
Eif4b
|
eukaryotic translation initiation factor 4B
|
|
chr11_-_63813083
Show fit
|
0.34 |
ENSMUST00000094103.4
|
Hs3st3b1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr13_-_69682696
Show fit
|
0.33 |
ENSMUST00000198607.5
|
Tent4a
|
terminal nucleotidyltransferase 4A
|
|
chr6_+_29348068
Show fit
|
0.31 |
ENSMUST00000173216.8
ENSMUST00000173694.5
ENSMUST00000172974.8
ENSMUST00000031779.17
ENSMUST00000090481.14
|
Calu
|
calumenin
|
|
chr7_+_46495256
Show fit
|
0.30 |
ENSMUST00000048209.16
ENSMUST00000210815.2
ENSMUST00000125862.8
ENSMUST00000210968.2
ENSMUST00000092621.12
ENSMUST00000210467.2
|
Ldha
|
lactate dehydrogenase A
|
|
chr16_+_36695479
Show fit
|
0.28 |
ENSMUST00000023534.7
ENSMUST00000114812.9
ENSMUST00000134616.8
|
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1
|
|
chr11_+_97206542
Show fit
|
0.26 |
ENSMUST00000019026.10
ENSMUST00000132168.2
|
Mrpl45
|
mitochondrial ribosomal protein L45
|
|
chr1_+_175708341
Show fit
|
0.25 |
ENSMUST00000195196.6
ENSMUST00000194306.6
ENSMUST00000193822.6
|
Exo1
|
exonuclease 1
|
|
chr4_-_126096551
Show fit
|
0.23 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3
|
|
chr7_+_44145987
Show fit
|
0.23 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1
|
|
chr11_-_113641980
Show fit
|
0.23 |
ENSMUST00000153453.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4
|
|
chr18_-_53877579
Show fit
|
0.23 |
ENSMUST00000049811.8
ENSMUST00000237062.2
|
Cep120
|
centrosomal protein 120
|
|
chr2_-_26012751
Show fit
|
0.22 |
ENSMUST00000140993.2
ENSMUST00000028300.6
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing
|
|
chr16_-_87229485
Show fit
|
0.22 |
ENSMUST00000039449.9
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1
|
|
chr15_-_81284244
Show fit
|
0.22 |
ENSMUST00000172107.8
ENSMUST00000169204.2
ENSMUST00000163382.2
|
St13
|
suppression of tumorigenicity 13
|
|
chr17_+_29768757
Show fit
|
0.22 |
ENSMUST00000048677.9
ENSMUST00000150388.3
|
Tbc1d22b
Gm28052
|
TBC1 domain family, member 22B
predicted gene, 28052
|
|
chr5_-_122917341
Show fit
|
0.21 |
ENSMUST00000198257.5
ENSMUST00000199599.2
ENSMUST00000196742.2
ENSMUST00000200109.5
ENSMUST00000111668.8
|
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr14_+_31807760
Show fit
|
0.21 |
ENSMUST00000170600.8
ENSMUST00000168986.7
ENSMUST00000169649.2
|
Oxnad1
|
oxidoreductase NAD-binding domain containing 1
|
|
chr15_-_79430942
Show fit
|
0.20 |
ENSMUST00000054014.9
ENSMUST00000229877.2
|
Ddx17
|
DEAD box helicase 17
|
|
chr19_+_46064409
Show fit
|
0.20 |
ENSMUST00000223728.2
ENSMUST00000235620.2
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr7_+_5023552
Show fit
|
0.20 |
ENSMUST00000208728.2
ENSMUST00000085427.6
|
Ccdc106
Zfp865
|
coiled-coil domain containing 106
zinc finger protein 865
|
|
chr16_+_49519561
Show fit
|
0.20 |
ENSMUST00000046777.11
ENSMUST00000142682.9
|
Ift57
|
intraflagellar transport 57
|
|
chrX_+_149371219
Show fit
|
0.19 |
ENSMUST00000153221.8
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr9_-_48406564
Show fit
|
0.17 |
ENSMUST00000213276.2
ENSMUST00000170000.4
|
Rbm7
|
RNA binding motif protein 7
|
|
chr11_+_22940599
Show fit
|
0.17 |
ENSMUST00000020562.5
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta)
|
|
chr19_+_46064302
Show fit
|
0.16 |
ENSMUST00000165017.2
ENSMUST00000223741.2
ENSMUST00000225780.2
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr9_+_48406706
Show fit
|
0.16 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617
|
|
chr7_+_44146012
Show fit
|
0.16 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1
|
|
chr17_+_84819260
Show fit
|
0.16 |
ENSMUST00000047206.7
|
Plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr5_-_113910571
Show fit
|
0.15 |
ENSMUST00000019118.8
|
Sart3
|
squamous cell carcinoma antigen recognized by T cells 3
|
|
chr2_+_30127692
Show fit
|
0.15 |
ENSMUST00000113654.8
ENSMUST00000095078.3
|
Lrrc8a
|
leucine rich repeat containing 8A VRAC subunit A
|
|
chr11_+_22940519
Show fit
|
0.15 |
ENSMUST00000173867.8
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta)
|
|
chr5_-_130053120
Show fit
|
0.15 |
ENSMUST00000161640.8
ENSMUST00000161884.2
ENSMUST00000161094.8
|
Asl
|
argininosuccinate lyase
|
|
chr15_+_98972850
Show fit
|
0.14 |
ENSMUST00000039665.8
|
Troap
|
trophinin associated protein
|
|
chr15_+_86098660
Show fit
|
0.13 |
ENSMUST00000063414.9
|
Tbc1d22a
|
TBC1 domain family, member 22a
|
|
chr8_-_25506916
Show fit
|
0.12 |
ENSMUST00000084035.12
ENSMUST00000208247.3
|
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma)
|
|
chr19_+_46064437
Show fit
|
0.12 |
ENSMUST00000223683.2
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr7_+_30021165
Show fit
|
0.11 |
ENSMUST00000098585.4
|
E130208F15Rik
|
RIKEN cDNA E130208F15 gene
|
|
chr17_-_56933872
Show fit
|
0.11 |
ENSMUST00000047226.10
|
Lonp1
|
lon peptidase 1, mitochondrial
|
|
chr10_-_78248771
Show fit
|
0.11 |
ENSMUST00000062678.11
|
Rrp1
|
ribosomal RNA processing 1
|
|
chr11_-_86435579
Show fit
|
0.10 |
ENSMUST00000138810.3
ENSMUST00000058286.9
ENSMUST00000154617.8
|
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1
|
|
chr11_-_85125889
Show fit
|
0.10 |
ENSMUST00000018625.10
|
Appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr5_-_93354348
Show fit
|
0.10 |
ENSMUST00000058550.15
|
Ccni
|
cyclin I
|
|
chr8_-_25506756
Show fit
|
0.10 |
ENSMUST00000084032.6
ENSMUST00000207132.2
|
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma)
|
|
chr6_-_128401832
Show fit
|
0.09 |
ENSMUST00000203374.3
ENSMUST00000142615.4
ENSMUST00000001559.11
|
Itfg2
|
integrin alpha FG-GAP repeat containing 2
|
|
chr17_+_23898633
Show fit
|
0.09 |
ENSMUST00000233364.2
|
Cldn6
|
claudin 6
|
|
chr19_-_24454720
Show fit
|
0.09 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A
|
|
chrX_+_111221031
Show fit
|
0.09 |
ENSMUST00000026599.10
ENSMUST00000113415.2
|
Apool
|
apolipoprotein O-like
|
|
chr16_+_34842764
Show fit
|
0.08 |
ENSMUST00000061156.10
|
Hacd2
|
3-hydroxyacyl-CoA dehydratase 2
|
|
chr7_+_5023375
Show fit
|
0.08 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865
|
|
chr11_-_113642135
Show fit
|
0.08 |
ENSMUST00000106616.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4
|
|
chr19_-_44124252
Show fit
|
0.06 |
ENSMUST00000026218.7
|
Cwf19l1
|
CWF19-like 1, cell cycle control (S. pombe)
|
|
chr17_+_85397980
Show fit
|
0.06 |
ENSMUST00000095188.7
|
Camkmt
|
calmodulin-lysine N-methyltransferase
|
|
chr7_+_101859542
Show fit
|
0.05 |
ENSMUST00000140631.2
ENSMUST00000120879.8
ENSMUST00000146996.8
|
Pgap2
|
post-GPI attachment to proteins 2
|
|
chr11_-_6576030
Show fit
|
0.05 |
ENSMUST00000000394.14
ENSMUST00000189268.7
ENSMUST00000136682.8
|
Tbrg4
|
transforming growth factor beta regulated gene 4
|
|
chr17_+_21165573
Show fit
|
0.04 |
ENSMUST00000007708.14
|
Ppp2r1a
|
protein phosphatase 2, regulatory subunit A, alpha
|
|
chr11_+_51153941
Show fit
|
0.02 |
ENSMUST00000093132.13
ENSMUST00000109113.8
|
Clk4
|
CDC like kinase 4
|
|
chr12_+_113104085
Show fit
|
0.01 |
ENSMUST00000200380.5
|
Crip2
|
cysteine rich protein 2
|
|
chr7_-_83533497
Show fit
|
0.00 |
ENSMUST00000094216.5
|
Tlnrd1
|
talin rod domain containing 1
|