Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
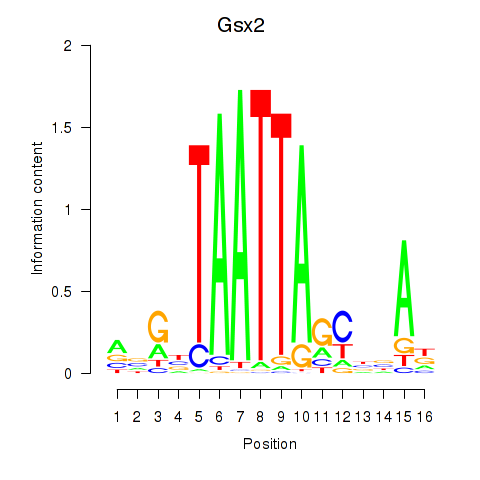
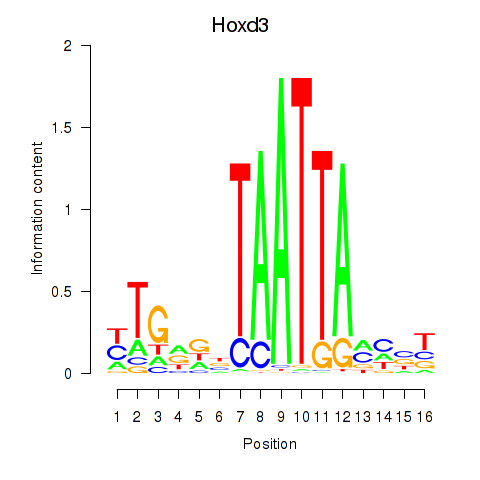
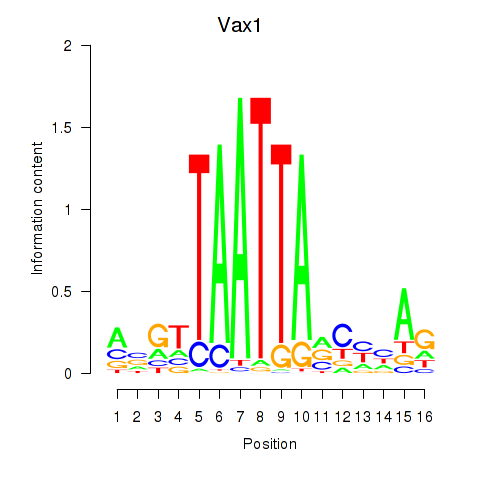
Results for Gsx2_Hoxd3_Vax1
Z-value: 0.66
Transcription factors associated with Gsx2_Hoxd3_Vax1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gsx2
|
ENSMUSG00000035946.8 | GS homeobox 2 |
|
Hoxd3
|
ENSMUSG00000079277.10 | homeobox D3 |
|
Vax1
|
ENSMUSG00000006270.8 | ventral anterior homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd3 | mm39_v1_chr2_+_74542255_74542283 | -0.06 | 7.3e-01 | Click! |
| Vax1 | mm39_v1_chr19_-_59158488_59158488 | 0.03 | 8.8e-01 | Click! |
Activity profile of Gsx2_Hoxd3_Vax1 motif
Sorted Z-values of Gsx2_Hoxd3_Vax1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_130754413 | 8.17 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr10_+_127734384 | 4.60 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr17_+_85335775 | 3.18 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr7_-_12829100 | 2.35 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr7_-_48493388 | 2.07 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr18_+_56565188 | 1.77 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr19_-_39637489 | 1.33 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr3_-_131096792 | 1.28 |
ENSMUST00000200236.2
ENSMUST00000106337.7 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr4_-_14621805 | 1.13 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr10_+_62860094 | 1.10 |
ENSMUST00000124784.8
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr6_-_41752111 | 1.10 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr4_-_14621669 | 1.09 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_-_116084810 | 1.03 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr11_+_116734104 | 1.01 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr5_+_87814058 | 1.01 |
ENSMUST00000199506.5
ENSMUST00000197631.5 ENSMUST00000094641.9 |
Csn1s1
|
casein alpha s1 |
| chr9_+_121780054 | 0.97 |
ENSMUST00000043011.9
ENSMUST00000214536.3 ENSMUST00000215990.3 |
Gask1a
|
golgi associated kinase 1A |
| chr10_+_62860291 | 0.95 |
ENSMUST00000020262.5
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr6_-_125357756 | 0.95 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr7_+_51528788 | 0.84 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr10_+_127919142 | 0.83 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr18_+_12874390 | 0.80 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr1_+_75412574 | 0.79 |
ENSMUST00000037796.14
ENSMUST00000113584.8 ENSMUST00000145166.8 ENSMUST00000143730.8 ENSMUST00000133418.8 ENSMUST00000144874.8 ENSMUST00000140287.8 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr15_-_8739893 | 0.73 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr15_-_101801351 | 0.72 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr5_-_137015683 | 0.71 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chrM_+_14138 | 0.67 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr16_-_45544960 | 0.65 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr7_-_24423715 | 0.62 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr4_-_148236516 | 0.61 |
ENSMUST00000056965.12
ENSMUST00000168503.8 ENSMUST00000152098.8 |
Fbxo6
|
F-box protein 6 |
| chr18_+_46874970 | 0.60 |
ENSMUST00000224622.2
|
Ap3s1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr10_+_128173603 | 0.60 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr4_-_14621497 | 0.59 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr4_+_8690398 | 0.59 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr3_-_58729732 | 0.58 |
ENSMUST00000191233.4
|
Mindy4b-ps
|
MINDY lysine 48 deubiquitinase 4B, pseudogene |
| chr13_-_53627110 | 0.57 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chrX_-_142716085 | 0.56 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr4_-_129121676 | 0.55 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr14_-_48900192 | 0.54 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr5_+_104350475 | 0.53 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chrX_+_159551009 | 0.53 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chrX_+_106299484 | 0.53 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr19_-_5610628 | 0.52 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr18_+_12874368 | 0.50 |
ENSMUST00000235000.2
ENSMUST00000115857.9 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr15_+_98468885 | 0.49 |
ENSMUST00000023728.8
|
Tex49
|
testis expressed 49 |
| chr10_-_129965752 | 0.48 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr4_+_100336003 | 0.47 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr8_+_84262409 | 0.47 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr11_+_43046476 | 0.46 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr18_+_46874920 | 0.43 |
ENSMUST00000025357.9
ENSMUST00000225520.2 |
Ap3s1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr8_+_46111703 | 0.43 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr16_-_29363671 | 0.42 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chrX_+_159551171 | 0.42 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr10_+_116111441 | 0.41 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr5_+_137015873 | 0.40 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr5_+_67125902 | 0.40 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr15_-_8740218 | 0.40 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr11_-_65160810 | 0.40 |
ENSMUST00000108695.9
|
Myocd
|
myocardin |
| chrM_+_10167 | 0.39 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr9_+_39932760 | 0.39 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr1_+_75412646 | 0.39 |
ENSMUST00000131545.8
ENSMUST00000141124.2 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr10_+_75983285 | 0.39 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr5_-_23881353 | 0.39 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr15_+_25774070 | 0.38 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr2_+_69727563 | 0.38 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr15_+_92495007 | 0.38 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr8_+_94879235 | 0.38 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chr5_+_87148697 | 0.38 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr11_-_49004584 | 0.38 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr9_+_110306020 | 0.37 |
ENSMUST00000198858.5
|
Kif9
|
kinesin family member 9 |
| chr3_+_138019040 | 0.36 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr16_+_51851917 | 0.36 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr16_+_51851948 | 0.36 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chrM_-_14061 | 0.35 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr14_+_26414422 | 0.35 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr1_-_83016152 | 0.34 |
ENSMUST00000164473.2
ENSMUST00000045560.15 |
Slc19a3
|
solute carrier family 19, member 3 |
| chr9_+_40092216 | 0.34 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr12_+_52746158 | 0.34 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr9_+_32027335 | 0.34 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chrX_-_100129626 | 0.34 |
ENSMUST00000113710.8
|
Slc7a3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr10_+_115854118 | 0.34 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chrX_+_36675081 | 0.34 |
ENSMUST00000115179.4
|
Rhox2d
|
reproductive homeobox 2D |
| chr7_-_102540089 | 0.33 |
ENSMUST00000217024.2
|
Olfr569
|
olfactory receptor 569 |
| chr9_+_110306052 | 0.33 |
ENSMUST00000197248.5
ENSMUST00000061155.12 ENSMUST00000198043.5 ENSMUST00000084952.8 |
Kif9
|
kinesin family member 9 |
| chrX_+_36712105 | 0.32 |
ENSMUST00000072167.10
ENSMUST00000184746.2 |
Rhox2e
|
reproductive homeobox 2E |
| chr1_-_172964941 | 0.32 |
ENSMUST00000200689.5
|
Olfr1408
|
olfactory receptor 1408 |
| chrX_-_98514278 | 0.32 |
ENSMUST00000113797.4
ENSMUST00000113790.8 ENSMUST00000036354.7 ENSMUST00000167246.2 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chrX_-_158921370 | 0.32 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr11_+_73489420 | 0.32 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chrX_-_142716200 | 0.31 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr7_-_123099672 | 0.31 |
ENSMUST00000042470.14
ENSMUST00000128217.2 |
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr6_-_93769426 | 0.31 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr13_+_22508759 | 0.30 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr11_-_65160767 | 0.30 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr3_+_63148887 | 0.30 |
ENSMUST00000194324.6
|
Mme
|
membrane metallo endopeptidase |
| chr2_+_177760959 | 0.30 |
ENSMUST00000108916.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr9_-_50571080 | 0.30 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr2_+_69727599 | 0.30 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_+_111610658 | 0.30 |
ENSMUST00000054004.2
|
Olfr1302
|
olfactory receptor 1302 |
| chr4_-_42661893 | 0.29 |
ENSMUST00000108006.4
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr13_-_23041731 | 0.28 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr2_+_111327525 | 0.28 |
ENSMUST00000121345.4
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr5_-_143279378 | 0.27 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chrX_+_36508629 | 0.27 |
ENSMUST00000063340.12
ENSMUST00000173650.2 |
Rhox2a
|
reproductive homeobox 2A |
| chr7_-_106354591 | 0.27 |
ENSMUST00000214306.2
ENSMUST00000216255.2 |
Olfr698
|
olfactory receptor 698 |
| chr11_-_99412084 | 0.27 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr11_+_84020475 | 0.26 |
ENSMUST00000133811.3
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr19_-_39801188 | 0.26 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr5_-_3697806 | 0.26 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr7_+_43885573 | 0.26 |
ENSMUST00000223070.2
ENSMUST00000205530.2 |
Gm36864
|
predicted gene, 36864 |
| chr2_+_177760768 | 0.26 |
ENSMUST00000108917.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr2_-_164455520 | 0.26 |
ENSMUST00000094351.11
ENSMUST00000109338.2 |
Wfdc8
|
WAP four-disulfide core domain 8 |
| chr5_-_99091681 | 0.25 |
ENSMUST00000162619.8
ENSMUST00000162147.6 |
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr11_+_59197746 | 0.25 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr4_-_150998857 | 0.25 |
ENSMUST00000105675.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr4_-_114991174 | 0.24 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr11_+_114741948 | 0.24 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_164455438 | 0.24 |
ENSMUST00000094346.3
|
Wfdc6b
|
WAP four-disulfide core domain 6B |
| chr4_+_133280680 | 0.24 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr2_+_88505972 | 0.24 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr3_-_88204286 | 0.23 |
ENSMUST00000107556.10
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr18_+_59195354 | 0.23 |
ENSMUST00000165666.9
|
Minar2
|
membrane integral NOTCH2 associated receptor 2 |
| chr14_-_36820304 | 0.23 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr7_-_43885552 | 0.22 |
ENSMUST00000236952.2
|
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr13_-_103042554 | 0.22 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_-_99412162 | 0.21 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr3_+_103646682 | 0.21 |
ENSMUST00000106852.2
|
Gm10964
|
predicted gene 10964 |
| chr12_-_104439589 | 0.21 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr13_-_103042294 | 0.21 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_+_17622934 | 0.21 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr11_+_58062467 | 0.21 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chrX_+_113384297 | 0.21 |
ENSMUST00000133447.2
|
Klhl4
|
kelch-like 4 |
| chr1_-_69726384 | 0.21 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr7_-_25098122 | 0.21 |
ENSMUST00000105177.3
ENSMUST00000149349.2 |
Lipe
|
lipase, hormone sensitive |
| chr1_-_163552693 | 0.20 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr3_-_75177378 | 0.20 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chrX_-_74460137 | 0.20 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr9_+_27210500 | 0.20 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr8_+_36956345 | 0.20 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr5_+_27022355 | 0.20 |
ENSMUST00000071500.13
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr14_-_109151590 | 0.20 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr12_-_79343040 | 0.20 |
ENSMUST00000218377.2
ENSMUST00000021547.8 |
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr5_-_84565218 | 0.19 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr17_+_79919267 | 0.19 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr17_+_88748139 | 0.19 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chr11_-_59466995 | 0.19 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr10_-_128918779 | 0.19 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr11_+_100978103 | 0.19 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
| chr8_+_46111778 | 0.18 |
ENSMUST00000143820.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_-_56546088 | 0.18 |
ENSMUST00000203372.3
|
Pde1c
|
phosphodiesterase 1C |
| chr6_+_8948608 | 0.18 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr11_+_60956624 | 0.18 |
ENSMUST00000041944.9
ENSMUST00000108717.3 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr9_-_107749594 | 0.17 |
ENSMUST00000183035.2
|
Rbm6
|
RNA binding motif protein 6 |
| chr17_-_37430949 | 0.17 |
ENSMUST00000214994.2
ENSMUST00000216341.2 |
Olfr92
|
olfactory receptor 92 |
| chr17_+_93506590 | 0.17 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr2_+_110427643 | 0.17 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr13_-_43634695 | 0.17 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr13_+_22656093 | 0.17 |
ENSMUST00000226330.2
ENSMUST00000226965.2 |
Vmn1r201
|
vomeronasal 1 receptor 201 |
| chr2_-_152422220 | 0.17 |
ENSMUST00000053180.4
|
Defb19
|
defensin beta 19 |
| chr2_-_164013033 | 0.17 |
ENSMUST00000045196.4
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chr12_+_38831093 | 0.17 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr14_+_54082691 | 0.17 |
ENSMUST00000103674.6
|
Trav19
|
T cell receptor alpha variable 19 |
| chr6_+_134617903 | 0.16 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr8_-_87307294 | 0.16 |
ENSMUST00000131423.8
ENSMUST00000152438.2 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr9_-_112016966 | 0.16 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr2_-_164231015 | 0.16 |
ENSMUST00000167427.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr3_-_64473251 | 0.16 |
ENSMUST00000176481.9
|
Vmn2r6
|
vomeronasal 2, receptor 6 |
| chr3_+_103739877 | 0.16 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chrM_+_9870 | 0.16 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr7_-_106408041 | 0.15 |
ENSMUST00000214840.2
|
Olfr700
|
olfactory receptor 700 |
| chr12_+_38830812 | 0.15 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr7_-_43885522 | 0.15 |
ENSMUST00000206686.2
ENSMUST00000037220.5 |
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr8_-_122268693 | 0.15 |
ENSMUST00000034265.11
|
1700018B08Rik
|
RIKEN cDNA 1700018B08 gene |
| chr1_+_54289833 | 0.15 |
ENSMUST00000027128.11
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr7_+_19102423 | 0.15 |
ENSMUST00000132655.2
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr17_+_93506435 | 0.15 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr4_+_150999019 | 0.15 |
ENSMUST00000135169.8
|
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr7_-_8164653 | 0.15 |
ENSMUST00000168807.3
|
Vmn2r41
|
vomeronasal 2, receptor 41 |
| chr6_-_130314465 | 0.15 |
ENSMUST00000088017.5
ENSMUST00000111998.9 |
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3 |
| chr1_+_87983099 | 0.15 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chrX_+_143253677 | 0.15 |
ENSMUST00000178233.2
|
Trpc5os
|
transient receptor potential cation channel, subfamily C, member 5, opposite strand |
| chrX_+_36593720 | 0.15 |
ENSMUST00000115191.7
|
Rhox2b
|
reproductive homeobox 2B |
| chr6_-_123395075 | 0.15 |
ENSMUST00000172199.3
|
Vmn2r20
|
vomeronasal 2, receptor 20 |
| chr6_-_130106861 | 0.14 |
ENSMUST00000014476.6
|
Klra8
|
killer cell lectin-like receptor, subfamily A, member 8 |
| chr11_+_58549642 | 0.14 |
ENSMUST00000214392.2
|
Olfr322
|
olfactory receptor 322 |
| chr14_-_52704952 | 0.14 |
ENSMUST00000206520.3
|
Olfr1508
|
olfactory receptor 1508 |
| chr1_+_88234454 | 0.14 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr11_+_115225557 | 0.14 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr14_+_119092107 | 0.14 |
ENSMUST00000100314.4
|
Cldn10
|
claudin 10 |
| chr17_+_32877851 | 0.14 |
ENSMUST00000235086.2
|
Cyp4f40
|
cytochrome P450, family 4, subfamily f, polypeptide 40 |
| chr5_+_67125759 | 0.14 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr18_+_52748978 | 0.14 |
ENSMUST00000072666.4
ENSMUST00000209270.2 |
Zfp474
|
zinc finger protein 474 |
| chr2_+_76480606 | 0.14 |
ENSMUST00000099986.3
|
Pjvk
|
pejvakin |
| chr8_-_55177510 | 0.13 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chr9_-_99302205 | 0.13 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr8_-_13940234 | 0.13 |
ENSMUST00000033839.9
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr12_-_87742525 | 0.13 |
ENSMUST00000164517.3
|
Eif1ad19
|
eukaryotic translation initiation factor 1A domain containing 19 |
| chr7_-_25112256 | 0.13 |
ENSMUST00000200880.4
ENSMUST00000074040.4 |
Cxcl17
|
chemokine (C-X-C motif) ligand 17 |
| chr14_-_45626198 | 0.13 |
ENSMUST00000226590.2
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr3_+_53396120 | 0.13 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr16_+_91184661 | 0.13 |
ENSMUST00000139503.2
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsx2_Hoxd3_Vax1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.7 | 2.1 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.3 | 2.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.9 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.2 | 0.7 | GO:1904753 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.2 | 1.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 0.6 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.2 | 0.5 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 0.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.6 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.4 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.8 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.5 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.7 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.7 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.8 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.4 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.6 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.3 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.3 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.2 | GO:1903168 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.3 | GO:0015819 | lysine transport(GO:0015819) |
| 0.1 | 0.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.3 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0042376 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 1.0 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 1.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 1.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.7 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 1.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 3.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 1.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 3.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 1.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 2.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 4.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 1.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 2.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.6 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.7 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.3 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 0.2 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0019962 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0070330 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.0 | 0.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 3.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |