Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
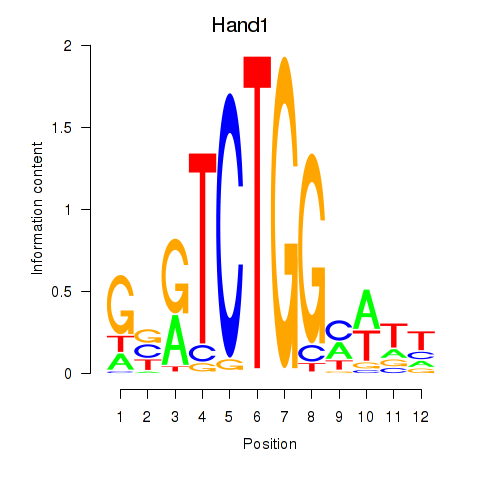
Results for Hand1
Z-value: 1.11
Transcription factors associated with Hand1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hand1
|
ENSMUSG00000037335.14 | heart and neural crest derivatives expressed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hand1 | mm39_v1_chr11_-_57722830_57722973 | 0.57 | 3.2e-04 | Click! |
Activity profile of Hand1 motif
Sorted Z-values of Hand1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_18618605 | 5.30 |
ENSMUST00000032573.8
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr10_+_75404482 | 4.80 |
ENSMUST00000134503.8
ENSMUST00000125770.8 ENSMUST00000128886.8 ENSMUST00000151212.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr10_+_127350820 | 3.77 |
ENSMUST00000035735.11
|
Ndufa4l2
|
Ndufa4, mitochondrial complex associated like 2 |
| chr4_-_60538151 | 3.67 |
ENSMUST00000098047.3
|
Mup10
|
major urinary protein 10 |
| chr4_-_60222580 | 3.57 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr10_+_75404464 | 3.34 |
ENSMUST00000145928.8
ENSMUST00000131565.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr2_-_120867529 | 3.30 |
ENSMUST00000102490.10
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr4_-_61592331 | 3.30 |
ENSMUST00000098040.4
|
Mup18
|
major urinary protein 18 |
| chr7_+_142559375 | 3.30 |
ENSMUST00000075172.12
ENSMUST00000105923.8 |
Tspan32
|
tetraspanin 32 |
| chr11_+_32226893 | 2.89 |
ENSMUST00000145569.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr4_-_60070411 | 2.83 |
ENSMUST00000079697.10
ENSMUST00000125282.2 ENSMUST00000166098.8 |
Mup7
|
major urinary protein 7 |
| chr14_-_54949596 | 2.66 |
ENSMUST00000064290.8
|
Cebpe
|
CCAAT/enhancer binding protein (C/EBP), epsilon |
| chr2_-_120867232 | 2.65 |
ENSMUST00000023987.6
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr7_+_24069680 | 2.64 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr7_+_142558783 | 2.61 |
ENSMUST00000009396.13
|
Tspan32
|
tetraspanin 32 |
| chr4_-_60377932 | 2.53 |
ENSMUST00000107506.9
ENSMUST00000122381.8 ENSMUST00000118759.8 ENSMUST00000132829.3 |
Mup9
|
major urinary protein 9 |
| chr7_+_142558837 | 2.38 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chr8_-_86091946 | 2.32 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr4_-_60618357 | 2.30 |
ENSMUST00000084544.5
ENSMUST00000098046.10 |
Mup11
|
major urinary protein 11 |
| chr13_-_37233179 | 2.29 |
ENSMUST00000037491.11
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr8_-_86091970 | 2.28 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr15_+_79232137 | 2.08 |
ENSMUST00000163691.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr5_-_124492734 | 2.07 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr17_-_26314461 | 2.02 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr4_+_114945905 | 1.95 |
ENSMUST00000171877.8
ENSMUST00000177647.8 ENSMUST00000106548.9 ENSMUST00000030488.3 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr1_-_131066004 | 1.95 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr17_-_26314438 | 1.92 |
ENSMUST00000236547.2
|
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr17_+_47906985 | 1.90 |
ENSMUST00000182539.8
|
Ccnd3
|
cyclin D3 |
| chr4_-_154110324 | 1.86 |
ENSMUST00000130175.8
ENSMUST00000182151.8 |
Smim1
|
small integral membrane protein 1 |
| chr7_+_16515265 | 1.84 |
ENSMUST00000108496.9
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr1_+_91294133 | 1.79 |
ENSMUST00000086861.12
|
Erfe
|
erythroferrone |
| chr6_-_60806810 | 1.69 |
ENSMUST00000163779.8
|
Snca
|
synuclein, alpha |
| chr6_-_60805873 | 1.69 |
ENSMUST00000114268.5
|
Snca
|
synuclein, alpha |
| chr15_+_79232591 | 1.68 |
ENSMUST00000229285.2
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr13_+_21679387 | 1.60 |
ENSMUST00000104942.2
|
AK157302
|
cDNA sequence AK157302 |
| chr14_-_36832044 | 1.60 |
ENSMUST00000179488.3
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr2_+_109522781 | 1.60 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr11_-_69838971 | 1.60 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr9_+_123195986 | 1.56 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr17_+_28988354 | 1.53 |
ENSMUST00000233109.2
ENSMUST00000004986.14 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr11_+_120653613 | 1.46 |
ENSMUST00000105046.4
|
Hmga1b
|
high mobility group AT-hook 1B |
| chr15_-_81845050 | 1.44 |
ENSMUST00000071462.7
ENSMUST00000023112.12 |
Pmm1
|
phosphomannomutase 1 |
| chr9_+_21176582 | 1.44 |
ENSMUST00000065005.5
|
Atg4d
|
autophagy related 4D, cysteine peptidase |
| chr17_+_28988271 | 1.43 |
ENSMUST00000233984.2
ENSMUST00000233460.2 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr11_-_102815910 | 1.43 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr1_+_134109888 | 1.40 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr7_-_141241632 | 1.40 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr11_+_97306353 | 1.39 |
ENSMUST00000121799.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr15_+_79982033 | 1.38 |
ENSMUST00000143928.2
|
Syngr1
|
synaptogyrin 1 |
| chr8_+_85449632 | 1.35 |
ENSMUST00000098571.5
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr4_+_109533753 | 1.33 |
ENSMUST00000102724.5
|
Faf1
|
Fas-associated factor 1 |
| chr17_+_56347424 | 1.33 |
ENSMUST00000002914.10
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr17_+_48761916 | 1.23 |
ENSMUST00000074574.13
|
Unc5cl
|
unc-5 family C-terminal like |
| chr4_-_43499608 | 1.23 |
ENSMUST00000136005.3
ENSMUST00000054538.13 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr10_-_128237087 | 1.19 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr4_+_120896582 | 1.19 |
ENSMUST00000030372.6
|
Col9a2
|
collagen, type IX, alpha 2 |
| chr18_-_74340885 | 1.18 |
ENSMUST00000177604.2
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr8_-_65489834 | 1.18 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr6_-_126916487 | 1.16 |
ENSMUST00000144954.5
ENSMUST00000112220.8 ENSMUST00000112221.8 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr7_-_4728081 | 1.15 |
ENSMUST00000086363.5
ENSMUST00000086364.11 |
Tmem150b
|
transmembrane protein 150B |
| chr5_-_77457895 | 1.15 |
ENSMUST00000047860.9
|
Noa1
|
nitric oxide associated 1 |
| chr2_-_32271921 | 1.14 |
ENSMUST00000048792.5
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr11_+_69805005 | 1.14 |
ENSMUST00000057884.6
|
Gps2
|
G protein pathway suppressor 2 |
| chr15_-_81845019 | 1.12 |
ENSMUST00000230229.2
|
Pmm1
|
phosphomannomutase 1 |
| chr8_+_57964921 | 1.12 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr18_-_74340842 | 1.10 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr11_+_4186391 | 1.09 |
ENSMUST00000075221.3
|
Osm
|
oncostatin M |
| chr3_+_96069271 | 1.09 |
ENSMUST00000054356.16
|
Mtmr11
|
myotubularin related protein 11 |
| chr17_-_74602469 | 1.08 |
ENSMUST00000233144.2
|
Memo1
|
mediator of cell motility 1 |
| chr7_-_30738471 | 1.05 |
ENSMUST00000162250.8
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr7_+_28488380 | 1.04 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr11_-_3454766 | 1.04 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr17_+_33774681 | 1.02 |
ENSMUST00000087605.13
ENSMUST00000174695.2 |
Myo1f
|
myosin IF |
| chr9_+_106083988 | 1.00 |
ENSMUST00000188650.2
|
Twf2
|
twinfilin actin binding protein 2 |
| chr7_+_130633776 | 0.99 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr8_-_65489791 | 0.98 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr1_-_120197979 | 0.98 |
ENSMUST00000112639.8
|
Steap3
|
STEAP family member 3 |
| chr5_-_138170644 | 0.98 |
ENSMUST00000000505.16
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr19_-_3979723 | 0.98 |
ENSMUST00000051803.8
|
Aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr12_-_91712783 | 0.97 |
ENSMUST00000166967.2
|
Ston2
|
stonin 2 |
| chr10_+_80662490 | 0.96 |
ENSMUST00000060987.15
ENSMUST00000177850.8 ENSMUST00000180036.8 ENSMUST00000179172.8 |
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr14_-_79718890 | 0.95 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr8_-_89362745 | 0.94 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr8_+_57964956 | 0.93 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr13_-_115226666 | 0.93 |
ENSMUST00000109226.5
|
Pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr2_+_132528851 | 0.92 |
ENSMUST00000061891.11
|
Shld1
|
shieldin complex subunit 1 |
| chr5_-_113968483 | 0.92 |
ENSMUST00000100874.6
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr5_+_35156389 | 0.88 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr4_-_117740624 | 0.87 |
ENSMUST00000030266.12
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr4_-_45084564 | 0.87 |
ENSMUST00000052236.13
|
Fbxo10
|
F-box protein 10 |
| chr18_+_67476664 | 0.86 |
ENSMUST00000025404.10
|
Cidea
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| chr3_-_90297187 | 0.83 |
ENSMUST00000029541.12
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr1_-_192883642 | 0.80 |
ENSMUST00000192020.6
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr7_-_127593003 | 0.80 |
ENSMUST00000033056.5
|
Pycard
|
PYD and CARD domain containing |
| chr19_-_4213347 | 0.79 |
ENSMUST00000025749.15
|
Rps6kb2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr1_-_192880260 | 0.79 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr7_-_30144933 | 0.79 |
ENSMUST00000006828.9
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr6_-_41613322 | 0.79 |
ENSMUST00000031902.7
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr4_+_119494901 | 0.78 |
ENSMUST00000024015.3
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr19_-_42074777 | 0.78 |
ENSMUST00000051772.10
|
Morn4
|
MORN repeat containing 4 |
| chr1_-_192883743 | 0.78 |
ENSMUST00000043550.11
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr1_+_163889713 | 0.77 |
ENSMUST00000097491.10
|
Sell
|
selectin, lymphocyte |
| chr8_-_106553822 | 0.77 |
ENSMUST00000239468.2
ENSMUST00000041400.6 |
Ranbp10
|
RAN binding protein 10 |
| chr9_+_53678801 | 0.75 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr14_-_51295099 | 0.75 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr11_-_58829738 | 0.74 |
ENSMUST00000094151.6
|
Rnf187
|
ring finger protein 187 |
| chr7_-_141614758 | 0.74 |
ENSMUST00000211000.2
ENSMUST00000209725.2 ENSMUST00000084418.4 |
Mob2
|
MOB kinase activator 2 |
| chr19_-_6178171 | 0.73 |
ENSMUST00000154601.8
ENSMUST00000138931.3 |
Snx15
|
sorting nexin 15 |
| chr15_+_97990431 | 0.71 |
ENSMUST00000229280.2
ENSMUST00000163507.8 ENSMUST00000230445.2 |
Pfkm
|
phosphofructokinase, muscle |
| chr2_-_32271833 | 0.70 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr14_+_54713557 | 0.70 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr4_+_130640436 | 0.68 |
ENSMUST00000151698.8
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr12_-_4283926 | 0.68 |
ENSMUST00000111169.10
ENSMUST00000020981.12 |
Cenpo
|
centromere protein O |
| chr13_+_21364330 | 0.68 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr2_-_126460575 | 0.67 |
ENSMUST00000028838.5
|
Hdc
|
histidine decarboxylase |
| chr4_+_134847949 | 0.67 |
ENSMUST00000056977.14
|
Runx3
|
runt related transcription factor 3 |
| chr5_-_138170077 | 0.67 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_124490296 | 0.66 |
ENSMUST00000111472.6
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr2_+_14878480 | 0.66 |
ENSMUST00000114719.7
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr11_-_114825089 | 0.66 |
ENSMUST00000149663.4
ENSMUST00000239005.2 ENSMUST00000106581.5 |
Cd300lb
|
CD300 molecule like family member B |
| chr8_-_108151661 | 0.65 |
ENSMUST00000003946.9
|
Nob1
|
NIN1/RPN12 binding protein 1 homolog |
| chr2_+_110427643 | 0.65 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chrX_-_74174608 | 0.65 |
ENSMUST00000033775.9
|
Mpp1
|
membrane protein, palmitoylated |
| chr17_+_34808772 | 0.64 |
ENSMUST00000038244.15
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr7_-_30119227 | 0.63 |
ENSMUST00000208740.2
ENSMUST00000075062.5 |
Hcst
|
hematopoietic cell signal transducer |
| chr19_+_4644365 | 0.63 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr7_-_19338349 | 0.63 |
ENSMUST00000086041.7
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr4_-_117744476 | 0.61 |
ENSMUST00000132073.2
ENSMUST00000150204.8 ENSMUST00000147845.2 ENSMUST00000036380.14 ENSMUST00000136596.2 |
Atp6v0b
|
ATPase, H+ transporting, lysosomal V0 subunit B |
| chr19_-_53020531 | 0.61 |
ENSMUST00000236008.2
ENSMUST00000237294.2 |
Xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr15_+_44320918 | 0.60 |
ENSMUST00000038336.12
ENSMUST00000166957.2 ENSMUST00000209244.2 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr1_+_74414354 | 0.60 |
ENSMUST00000187516.7
ENSMUST00000027368.6 |
Slc11a1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
| chr6_+_92846338 | 0.60 |
ENSMUST00000113434.2
|
Gm15737
|
predicted gene 15737 |
| chr14_+_65187485 | 0.59 |
ENSMUST00000043914.8
ENSMUST00000239450.2 |
Ints9
|
integrator complex subunit 9 |
| chr13_+_21364069 | 0.58 |
ENSMUST00000021761.13
|
Trim27
|
tripartite motif-containing 27 |
| chr8_+_55024446 | 0.58 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr5_-_65248962 | 0.58 |
ENSMUST00000212080.2
|
Tmem156
|
transmembrane protein 156 |
| chr10_+_33780993 | 0.58 |
ENSMUST00000169670.8
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr18_+_50112580 | 0.57 |
ENSMUST00000179937.2
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr7_-_126651120 | 0.57 |
ENSMUST00000051122.7
|
Zg16
|
zymogen granule protein 16 |
| chr19_-_4171536 | 0.56 |
ENSMUST00000025767.14
|
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chr2_+_149672760 | 0.56 |
ENSMUST00000109934.2
ENSMUST00000140870.8 |
Syndig1
|
synapse differentiation inducing 1 |
| chr9_+_106158549 | 0.55 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A |
| chr12_-_32258604 | 0.55 |
ENSMUST00000053215.14
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr2_+_149672708 | 0.55 |
ENSMUST00000109935.8
|
Syndig1
|
synapse differentiation inducing 1 |
| chr12_-_76842263 | 0.54 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr19_+_47167259 | 0.54 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr14_+_79753055 | 0.53 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr12_-_32258331 | 0.52 |
ENSMUST00000220366.2
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr18_-_64649620 | 0.51 |
ENSMUST00000025483.11
ENSMUST00000237400.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr19_+_45352173 | 0.51 |
ENSMUST00000223764.2
ENSMUST00000065601.13 ENSMUST00000224102.2 ENSMUST00000111936.4 |
Btrc
|
beta-transducin repeat containing protein |
| chr5_+_43976218 | 0.50 |
ENSMUST00000101237.8
|
Bst1
|
bone marrow stromal cell antigen 1 |
| chr9_+_69360902 | 0.50 |
ENSMUST00000034756.15
ENSMUST00000123470.8 |
Anxa2
|
annexin A2 |
| chr12_-_8589545 | 0.50 |
ENSMUST00000095863.10
ENSMUST00000165657.3 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr17_-_27842412 | 0.50 |
ENSMUST00000025050.13
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr2_-_113678945 | 0.49 |
ENSMUST00000110949.9
|
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr2_+_130969200 | 0.48 |
ENSMUST00000099349.10
ENSMUST00000100763.9 |
Hspa12b
|
heat shock protein 12B |
| chr2_+_118493713 | 0.48 |
ENSMUST00000099557.10
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr12_-_76869282 | 0.48 |
ENSMUST00000021459.14
|
Rab15
|
RAB15, member RAS oncogene family |
| chr1_+_75483721 | 0.48 |
ENSMUST00000037330.5
|
Inha
|
inhibin alpha |
| chrX_+_162922317 | 0.47 |
ENSMUST00000112271.10
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr12_-_32258469 | 0.47 |
ENSMUST00000085469.6
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr16_+_36648728 | 0.47 |
ENSMUST00000114819.8
ENSMUST00000023535.4 |
Iqcb1
|
IQ calmodulin-binding motif containing 1 |
| chr5_-_114828967 | 0.47 |
ENSMUST00000112212.2
ENSMUST00000112214.8 |
Gltp
|
glycolipid transfer protein |
| chr4_+_155887978 | 0.47 |
ENSMUST00000030942.13
ENSMUST00000185148.8 ENSMUST00000130188.2 |
Mrpl20
|
mitochondrial ribosomal protein L20 |
| chr3_-_106313406 | 0.47 |
ENSMUST00000029510.9
|
Chil6
|
chitinase-like 6 |
| chr14_+_54713703 | 0.47 |
ENSMUST00000164697.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr11_-_69786324 | 0.47 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr2_+_149672814 | 0.46 |
ENSMUST00000137280.2
ENSMUST00000149705.2 |
Syndig1
|
synapse differentiation inducing 1 |
| chr1_+_135656885 | 0.45 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr13_+_120761861 | 0.45 |
ENSMUST00000225029.2
|
BC147527
|
cDNA sequence BC147527 |
| chr2_-_152185901 | 0.45 |
ENSMUST00000040312.7
|
Trib3
|
tribbles pseudokinase 3 |
| chr11_-_54853729 | 0.44 |
ENSMUST00000108885.8
ENSMUST00000102730.9 ENSMUST00000018482.13 ENSMUST00000108886.8 ENSMUST00000102731.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr7_-_119078472 | 0.44 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr10_+_83558729 | 0.44 |
ENSMUST00000150459.3
|
1500009L16Rik
|
RIKEN cDNA 1500009L16 gene |
| chr2_+_158344532 | 0.44 |
ENSMUST00000059889.4
|
Adig
|
adipogenin |
| chr9_+_56772301 | 0.44 |
ENSMUST00000035661.7
|
Cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr18_-_52662728 | 0.43 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
| chr2_-_167852538 | 0.42 |
ENSMUST00000099073.3
|
Ripor3
|
RIPOR family member 3 |
| chr19_-_4213284 | 0.41 |
ENSMUST00000118483.2
ENSMUST00000155303.3 |
Rps6kb2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr5_+_20112500 | 0.41 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_+_120411588 | 0.41 |
ENSMUST00000208668.2
|
Gm5737
|
predicted gene 5737 |
| chr3_+_99161070 | 0.40 |
ENSMUST00000029462.10
|
Tbx15
|
T-box 15 |
| chr15_+_51740825 | 0.40 |
ENSMUST00000137116.3
ENSMUST00000161651.2 ENSMUST00000059599.10 |
Utp23
|
UTP23 small subunit processome component |
| chr10_-_49659355 | 0.40 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr4_-_41731142 | 0.40 |
ENSMUST00000171251.8
|
Arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr1_-_130815144 | 0.40 |
ENSMUST00000121040.8
|
Il24
|
interleukin 24 |
| chr19_-_47680528 | 0.39 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr7_-_30262659 | 0.39 |
ENSMUST00000043898.8
|
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr15_-_101759212 | 0.39 |
ENSMUST00000023790.5
|
Krt1
|
keratin 1 |
| chr4_+_134042423 | 0.39 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr5_+_67125902 | 0.39 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr5_-_121045568 | 0.38 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr1_-_75168567 | 0.38 |
ENSMUST00000040689.15
ENSMUST00000189702.7 |
Atg9a
|
autophagy related 9A |
| chr7_-_30262337 | 0.38 |
ENSMUST00000207031.2
|
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr5_+_20112704 | 0.38 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_+_84966569 | 0.37 |
ENSMUST00000057019.9
|
Aplnr
|
apelin receptor |
| chr18_-_64649497 | 0.37 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr3_-_141874955 | 0.37 |
ENSMUST00000098568.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr6_-_68887922 | 0.36 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr6_-_128332789 | 0.36 |
ENSMUST00000001562.9
|
Tulp3
|
tubby-like protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hand1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 1.1 | 3.4 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 1.0 | 8.1 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.8 | 4.6 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.8 | 8.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.6 | 2.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.6 | 1.8 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.5 | 1.6 | GO:0061193 | taste bud development(GO:0061193) |
| 0.5 | 1.6 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.4 | 8.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.4 | 1.9 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.3 | 3.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 0.8 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.3 | 0.8 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 1.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 1.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 2.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 0.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 1.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 | 0.8 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.2 | 1.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 1.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 0.6 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.2 | 1.0 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.2 | 1.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.5 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.2 | 0.9 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 0.9 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.2 | 0.9 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.9 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 0.7 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.2 | 1.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 2.6 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 | 2.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 0.5 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.1 | 1.8 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.9 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.3 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.7 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 0.6 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.5 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.7 | GO:0001692 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) |
| 0.1 | 0.6 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 1.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.1 | 1.0 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 0.7 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.4 | GO:0072218 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.3 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.3 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 0.5 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 0.3 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.5 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.4 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.1 | 0.6 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 1.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 0.4 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 2.3 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.1 | 0.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 1.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 1.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 1.8 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.4 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.2 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.1 | 0.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 1.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.2 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.3 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 2.7 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 1.9 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 0.2 | GO:0010925 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.1 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.1 | 1.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.5 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.2 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.3 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.1 | 3.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 1.0 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.9 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.6 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 1.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.6 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 2.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.7 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 1.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 1.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.7 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 2.9 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.1 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.5 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.7 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.7 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.7 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 1.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.0 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.6 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 1.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.0 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:1901249 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.4 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.8 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.3 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.8 | 3.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 2.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 1.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 1.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.3 | 1.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 0.7 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.2 | 4.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.8 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 1.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 0.7 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.2 | 0.5 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.2 | 2.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.0 | 0.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 3.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 6.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 1.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.3 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.9 | 2.6 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.7 | 7.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.6 | 7.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.5 | 1.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 4.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 1.6 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.4 | 3.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 1.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.3 | 2.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 1.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 3.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 1.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 2.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 0.9 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 1.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 0.9 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.2 | 2.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.8 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 0.6 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.2 | 0.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 1.9 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 4.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.8 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.5 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 1.0 | GO:0008823 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.8 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 3.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 1.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.3 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 3.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.8 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.8 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.2 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.2 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.2 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 1.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.6 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 3.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.9 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.6 | GO:0003774 | motor activity(GO:0003774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 8.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 2.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 4.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 2.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 2.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |