Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
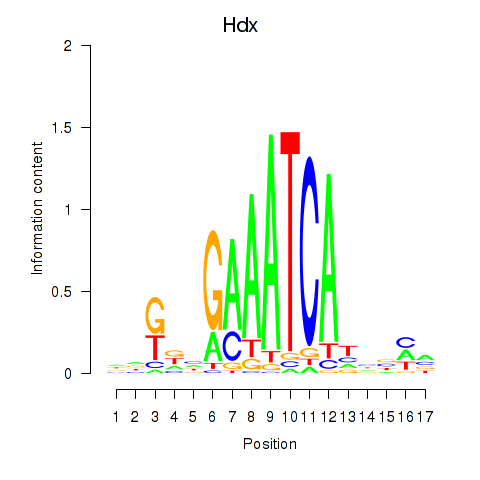
Results for Hdx
Z-value: 1.87
Transcription factors associated with Hdx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hdx
|
ENSMUSG00000034551.13 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hdx | mm39_v1_chrX_-_110606766_110606834 | -0.62 | 6.6e-05 | Click! |
Activity profile of Hdx motif
Sorted Z-values of Hdx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_61437704 | 28.35 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr4_-_62069046 | 21.34 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr19_+_20470056 | 17.76 |
ENSMUST00000225337.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr15_+_4756657 | 14.48 |
ENSMUST00000162585.8
|
C6
|
complement component 6 |
| chr19_+_20470114 | 14.24 |
ENSMUST00000225313.2
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr5_-_87240405 | 13.95 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr15_+_4756684 | 13.88 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr17_-_34962823 | 12.32 |
ENSMUST00000069507.9
|
C4b
|
complement component 4B (Chido blood group) |
| chr19_-_46661501 | 11.65 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr1_+_87998487 | 9.97 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr15_+_10177709 | 9.30 |
ENSMUST00000124470.8
|
Prlr
|
prolactin receptor |
| chr19_-_46661321 | 9.22 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr13_-_56696310 | 9.22 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr13_-_56696222 | 8.66 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr15_+_10249646 | 7.35 |
ENSMUST00000134410.8
|
Prlr
|
prolactin receptor |
| chr9_-_71075939 | 7.12 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr6_-_3968365 | 7.03 |
ENSMUST00000031674.11
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr1_-_140111138 | 6.32 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 5.95 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr7_-_5128936 | 5.38 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr16_+_22769822 | 5.22 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr4_+_155666933 | 5.22 |
ENSMUST00000105612.2
|
Nadk
|
NAD kinase |
| chr10_-_126956991 | 4.70 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr16_+_22769844 | 4.64 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr16_+_22926162 | 4.17 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr8_-_85500010 | 4.06 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr17_+_31652073 | 3.92 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr17_+_31652029 | 3.89 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr14_-_45715308 | 3.81 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr13_+_24511387 | 3.81 |
ENSMUST00000224953.2
ENSMUST00000050859.13 ENSMUST00000167746.8 ENSMUST00000224819.2 |
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr1_+_13738967 | 3.78 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr1_-_184543367 | 3.73 |
ENSMUST00000048462.13
ENSMUST00000110992.9 |
Mtarc1
|
mitochondrial amidoxime reducing component 1 |
| chr17_-_45997823 | 3.43 |
ENSMUST00000156254.8
|
Tmem63b
|
transmembrane protein 63b |
| chr13_-_96028412 | 3.40 |
ENSMUST00000068603.8
|
Iqgap2
|
IQ motif containing GTPase activating protein 2 |
| chr7_-_29935150 | 3.35 |
ENSMUST00000189482.2
|
Ovol3
|
ovo like zinc finger 3 |
| chr1_+_182392559 | 3.11 |
ENSMUST00000168514.7
|
Capn8
|
calpain 8 |
| chr3_-_154036180 | 3.06 |
ENSMUST00000177846.8
|
Lhx8
|
LIM homeobox protein 8 |
| chr17_+_3447465 | 3.05 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr6_-_87327885 | 3.03 |
ENSMUST00000032129.3
|
Gkn1
|
gastrokine 1 |
| chr2_+_69210775 | 2.99 |
ENSMUST00000063690.4
|
Dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_-_171122509 | 2.82 |
ENSMUST00000111302.4
ENSMUST00000080001.9 |
Ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr6_+_54244116 | 2.81 |
ENSMUST00000114402.9
|
Chn2
|
chimerin 2 |
| chr6_-_4086914 | 2.79 |
ENSMUST00000049166.5
|
Bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr18_-_35631914 | 2.78 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr14_+_53743184 | 2.75 |
ENSMUST00000103583.5
|
Trav10
|
T cell receptor alpha variable 10 |
| chr9_-_39918243 | 2.61 |
ENSMUST00000073932.4
|
Olfr980
|
olfactory receptor 980 |
| chr9_+_14187597 | 2.58 |
ENSMUST00000208222.2
|
Sesn3
|
sestrin 3 |
| chr2_-_121637505 | 2.51 |
ENSMUST00000138157.8
|
Frmd5
|
FERM domain containing 5 |
| chr3_+_62327089 | 2.49 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr4_+_89055359 | 2.32 |
ENSMUST00000058030.10
|
Mtap
|
methylthioadenosine phosphorylase |
| chr1_+_182392577 | 2.32 |
ENSMUST00000048941.14
|
Capn8
|
calpain 8 |
| chr2_-_121637469 | 2.28 |
ENSMUST00000110592.2
|
Frmd5
|
FERM domain containing 5 |
| chr13_+_41403317 | 2.21 |
ENSMUST00000165561.4
|
Smim13
|
small integral membrane protein 13 |
| chr6_+_41107047 | 2.20 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr3_+_122305819 | 2.17 |
ENSMUST00000199344.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr3_-_89152320 | 2.08 |
ENSMUST00000107464.8
ENSMUST00000090924.13 |
Trim46
|
tripartite motif-containing 46 |
| chr4_-_57300361 | 2.05 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr14_+_53995108 | 2.05 |
ENSMUST00000184905.2
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr11_+_97692738 | 2.04 |
ENSMUST00000127033.9
|
Lasp1
|
LIM and SH3 protein 1 |
| chr12_-_81615248 | 2.03 |
ENSMUST00000008582.4
|
Adam21
|
a disintegrin and metallopeptidase domain 21 |
| chr11_+_98337655 | 1.94 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr14_-_36690636 | 1.84 |
ENSMUST00000067700.13
|
Ccser2
|
coiled-coil serine rich 2 |
| chr18_-_39622295 | 1.79 |
ENSMUST00000131885.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr3_+_115801869 | 1.77 |
ENSMUST00000106502.2
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr5_-_146900548 | 1.72 |
ENSMUST00000125217.2
ENSMUST00000110564.8 ENSMUST00000066675.10 ENSMUST00000016654.9 ENSMUST00000110566.2 ENSMUST00000140526.2 |
Mtif3
|
mitochondrial translational initiation factor 3 |
| chr14_-_68893253 | 1.68 |
ENSMUST00000225767.3
ENSMUST00000111072.8 ENSMUST00000022642.6 ENSMUST00000224039.2 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr14_+_62901114 | 1.64 |
ENSMUST00000171692.2
|
Serpine3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr8_-_23196523 | 1.54 |
ENSMUST00000033939.13
ENSMUST00000063401.10 |
Ikbkb
|
inhibitor of kappaB kinase beta |
| chr9_+_27210500 | 1.52 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr11_-_69576363 | 1.52 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_-_155571279 | 1.48 |
ENSMUST00000040833.5
|
Edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr18_-_61533434 | 1.45 |
ENSMUST00000063307.6
ENSMUST00000075299.13 |
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta |
| chr11_-_50778270 | 1.43 |
ENSMUST00000050595.13
ENSMUST00000163301.8 ENSMUST00000109131.8 ENSMUST00000125749.2 |
Zfp454
|
zinc finger protein 454 |
| chr11_+_49176109 | 1.43 |
ENSMUST00000217275.2
ENSMUST00000214598.2 ENSMUST00000215861.2 ENSMUST00000214170.2 |
Olfr1392
|
olfactory receptor 1392 |
| chr15_-_98985698 | 1.42 |
ENSMUST00000064462.5
|
C1ql4
|
complement component 1, q subcomponent-like 4 |
| chr11_+_102175985 | 1.40 |
ENSMUST00000156326.2
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr18_-_72484126 | 1.40 |
ENSMUST00000114943.11
|
Dcc
|
deleted in colorectal carcinoma |
| chrX_+_106193060 | 1.39 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr16_+_14523696 | 1.35 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr17_-_45970238 | 1.34 |
ENSMUST00000120717.8
|
Capn11
|
calpain 11 |
| chr7_+_86564302 | 1.33 |
ENSMUST00000233033.2
ENSMUST00000170835.4 |
Vmn2r78
|
vomeronasal 2, receptor 78 |
| chr13_-_40882417 | 1.32 |
ENSMUST00000225180.2
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr11_-_69686756 | 1.31 |
ENSMUST00000045971.9
|
Chrnb1
|
cholinergic receptor, nicotinic, beta polypeptide 1 (muscle) |
| chr5_+_107585774 | 1.27 |
ENSMUST00000162298.4
ENSMUST00000094541.4 ENSMUST00000211896.2 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr15_+_100659622 | 1.25 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr11_+_43046476 | 1.25 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr5_+_107745562 | 1.24 |
ENSMUST00000112651.8
ENSMUST00000112654.8 ENSMUST00000065422.12 |
Rpap2
|
RNA polymerase II associated protein 2 |
| chr16_-_50252703 | 1.23 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr8_-_43760017 | 1.22 |
ENSMUST00000082120.5
|
Zfp42
|
zinc finger protein 42 |
| chr2_+_36575800 | 1.18 |
ENSMUST00000213258.2
|
Olfr346
|
olfactory receptor 346 |
| chr11_+_117123107 | 1.14 |
ENSMUST00000106354.9
|
Septin9
|
septin 9 |
| chr9_-_45115381 | 1.13 |
ENSMUST00000034599.15
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr14_+_53872276 | 1.13 |
ENSMUST00000103658.4
|
Trav13-2
|
T cell receptor alpha variable 13-2 |
| chr18_+_77877611 | 1.12 |
ENSMUST00000238172.2
|
Pstpip2
|
proline-serine-threonine phosphatase-interacting protein 2 |
| chr5_+_107745626 | 1.10 |
ENSMUST00000112655.8
|
Rpap2
|
RNA polymerase II associated protein 2 |
| chrX_+_106193167 | 1.10 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr1_+_37068387 | 1.05 |
ENSMUST00000067178.14
ENSMUST00000238500.2 |
Vwa3b
|
von Willebrand factor A domain containing 3B |
| chr17_-_40553176 | 1.04 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr12_+_89779178 | 1.03 |
ENSMUST00000238943.2
|
Nrxn3
|
neurexin III |
| chr7_+_19093665 | 1.03 |
ENSMUST00000140836.8
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chrX_-_111315519 | 1.02 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr9_+_67666705 | 1.01 |
ENSMUST00000171652.3
|
C2cd4b
|
C2 calcium-dependent domain containing 4B |
| chr4_-_114991174 | 1.01 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr1_+_37068429 | 1.01 |
ENSMUST00000162449.7
|
Vwa3b
|
von Willebrand factor A domain containing 3B |
| chr11_+_67477501 | 1.00 |
ENSMUST00000108680.2
|
Gas7
|
growth arrest specific 7 |
| chr11_-_99941369 | 1.00 |
ENSMUST00000007318.2
|
Krt31
|
keratin 31 |
| chr7_-_9575286 | 0.97 |
ENSMUST00000174433.2
|
Gm10302
|
predicted gene 10302 |
| chrX_-_20955370 | 0.97 |
ENSMUST00000040667.13
|
Zfp300
|
zinc finger protein 300 |
| chr2_-_17465410 | 0.96 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr8_+_110220614 | 0.94 |
ENSMUST00000034162.8
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr12_+_89779237 | 0.93 |
ENSMUST00000110133.9
ENSMUST00000110130.4 |
Nrxn3
|
neurexin III |
| chr2_+_86655007 | 0.92 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr1_-_179527845 | 0.91 |
ENSMUST00000223392.2
|
Kif28
|
kinesin family member 28 |
| chr18_-_72484082 | 0.90 |
ENSMUST00000073379.6
|
Dcc
|
deleted in colorectal carcinoma |
| chrX_+_142301666 | 0.90 |
ENSMUST00000134402.8
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr9_+_3015654 | 0.88 |
ENSMUST00000099050.4
|
Gm10720
|
predicted gene 10720 |
| chr5_-_129907878 | 0.87 |
ENSMUST00000026617.13
|
Phkg1
|
phosphorylase kinase gamma 1 |
| chr2_-_37249247 | 0.85 |
ENSMUST00000112940.8
ENSMUST00000009174.15 |
Pdcl
|
phosducin-like |
| chr13_+_13357291 | 0.85 |
ENSMUST00000021778.14
ENSMUST00000126540.8 ENSMUST00000151144.2 |
Prl2c5
|
prolactin family 2, subfamily c, member 5 |
| chr6_-_129308748 | 0.84 |
ENSMUST00000051283.8
|
Clec2m
|
C-type lectin domain family 2, member m |
| chr6_-_78355834 | 0.84 |
ENSMUST00000089667.8
ENSMUST00000167492.4 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr2_+_101716577 | 0.83 |
ENSMUST00000028584.8
|
Commd9
|
COMM domain containing 9 |
| chr4_-_57300750 | 0.75 |
ENSMUST00000151964.2
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr4_+_3938903 | 0.75 |
ENSMUST00000121210.8
ENSMUST00000121651.8 ENSMUST00000041122.11 ENSMUST00000120732.8 ENSMUST00000119307.8 ENSMUST00000123769.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr9_+_3025417 | 0.75 |
ENSMUST00000075573.7
|
Gm10717
|
predicted gene 10717 |
| chr6_-_143892814 | 0.71 |
ENSMUST00000124233.7
ENSMUST00000144289.4 ENSMUST00000111748.8 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr17_-_44416665 | 0.70 |
ENSMUST00000024757.14
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr5_-_26177661 | 0.70 |
ENSMUST00000200447.2
|
Gm21680
|
predicted gene, 21680 |
| chr5_-_86666408 | 0.70 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr5_-_107745404 | 0.69 |
ENSMUST00000124140.2
|
Glmn
|
glomulin, FKBP associated protein |
| chr5_-_109340065 | 0.69 |
ENSMUST00000053253.10
|
Vmn2r13
|
vomeronasal 2, receptor 13 |
| chr4_-_97472844 | 0.68 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr6_-_130170075 | 0.68 |
ENSMUST00000112032.8
ENSMUST00000071554.3 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr5_+_15824062 | 0.67 |
ENSMUST00000095006.9
|
Speer4d
|
spermatogenesis associated glutamate (E)-rich protein 4D |
| chr19_-_32038838 | 0.65 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr3_-_64417263 | 0.65 |
ENSMUST00000177184.9
|
Vmn2r5
|
vomeronasal 2, receptor 5 |
| chr11_-_67856457 | 0.64 |
ENSMUST00000021287.12
ENSMUST00000126766.2 |
Cfap52
|
cilia and flagella associated protein 52 |
| chr1_+_87254729 | 0.64 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr15_+_102862862 | 0.64 |
ENSMUST00000001701.4
|
Hoxc11
|
homeobox C11 |
| chr16_-_50151350 | 0.63 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chr2_-_151528259 | 0.63 |
ENSMUST00000149319.2
|
Tmem74bos
|
transmembrane 74B, opposite strand |
| chrX_+_106192510 | 0.63 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr13_-_67877228 | 0.61 |
ENSMUST00000073157.15
|
Zfp65
|
zinc finger protein 65 |
| chr1_+_179928709 | 0.61 |
ENSMUST00000133890.8
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr7_+_66489500 | 0.60 |
ENSMUST00000107478.9
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chrX_+_142301572 | 0.60 |
ENSMUST00000033640.14
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr9_+_3004457 | 0.59 |
ENSMUST00000178348.2
|
Gm11168
|
predicted gene 11168 |
| chr18_-_32082624 | 0.59 |
ENSMUST00000064016.6
|
Gpr17
|
G protein-coupled receptor 17 |
| chr17_+_38032699 | 0.59 |
ENSMUST00000207771.3
|
Olfr120
|
olfactory receptor 120 |
| chr3_-_144804784 | 0.58 |
ENSMUST00000040465.11
ENSMUST00000198993.2 |
Clca2
|
chloride channel accessory 2 |
| chr5_-_26310360 | 0.58 |
ENSMUST00000063524.3
|
5031410I06Rik
|
RIKEN cDNA 5031410I06 gene |
| chr5_-_26326419 | 0.58 |
ENSMUST00000088236.5
|
Gm10220
|
predicted gene 10220 |
| chr3_-_34135408 | 0.58 |
ENSMUST00000117223.4
ENSMUST00000120805.8 ENSMUST00000011029.12 ENSMUST00000108195.10 |
Dnajc19
|
DnaJ heat shock protein family (Hsp40) member C19 |
| chr5_-_26227914 | 0.58 |
ENSMUST00000072286.7
|
Gm5862
|
predicted gene 5862 |
| chr12_-_87742525 | 0.58 |
ENSMUST00000164517.3
|
Eif1ad19
|
eukaryotic translation initiation factor 1A domain containing 19 |
| chr5_-_26147379 | 0.57 |
ENSMUST00000196214.2
|
Gm21663
|
predicted gene, 21663 |
| chr5_-_26263579 | 0.57 |
ENSMUST00000095004.4
|
Gm7347
|
predicted gene 7347 |
| chr9_+_3037111 | 0.56 |
ENSMUST00000177969.2
|
Gm10715
|
predicted gene 10715 |
| chr6_+_30568366 | 0.56 |
ENSMUST00000049251.6
|
Cpa4
|
carboxypeptidase A4 |
| chr17_+_33651864 | 0.56 |
ENSMUST00000174088.3
|
Actl9
|
actin-like 9 |
| chr5_-_15681521 | 0.56 |
ENSMUST00000197995.2
|
Gm21149
|
predicted gene, 21149 |
| chr5_+_15782286 | 0.56 |
ENSMUST00000188807.4
|
Gm21083
|
predicted gene, 21083 |
| chr5_-_15919234 | 0.55 |
ENSMUST00000095005.12
ENSMUST00000179506.2 |
Speer4c
|
spermatogenesis associated glutamate (E)-rich protein 4C |
| chr11_+_81992662 | 0.54 |
ENSMUST00000000194.4
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr13_+_22737368 | 0.54 |
ENSMUST00000228195.2
|
Vmn1r204
|
vomeronasal 1 receptor 204 |
| chr13_-_67417132 | 0.54 |
ENSMUST00000045969.8
|
Zfp458
|
zinc finger protein 458 |
| chrX_-_8455485 | 0.54 |
ENSMUST00000089379.5
|
Ssxb3
|
synovial sarcoma, X member B3 |
| chrX_+_133088238 | 0.54 |
ENSMUST00000064476.5
|
Arl13a
|
ADP-ribosylation factor-like 13A |
| chr9_+_3018753 | 0.53 |
ENSMUST00000179272.2
|
Gm10719
|
predicted gene 10719 |
| chr5_+_26462689 | 0.52 |
ENSMUST00000074148.7
|
Gm7361
|
predicted gene 7361 |
| chr7_-_9687512 | 0.52 |
ENSMUST00000165611.2
|
Vmn2r48
|
vomeronasal 2, receptor 48 |
| chr5_-_15028949 | 0.51 |
ENSMUST00000096953.5
|
Gm10354
|
predicted gene 10354 |
| chr7_-_41708447 | 0.51 |
ENSMUST00000168489.3
ENSMUST00000233456.2 |
Vmn2r59
|
vomeronasal 2, receptor 59 |
| chr8_-_84169071 | 0.51 |
ENSMUST00000212449.2
|
Scoc
|
short coiled-coil protein |
| chr5_-_26244556 | 0.50 |
ENSMUST00000079447.4
|
Speer4a
|
spermatogenesis associated glutamate (E)-rich protein 4A |
| chr16_-_37360097 | 0.50 |
ENSMUST00000023525.9
|
Gtf2e1
|
general transcription factor II E, polypeptide 1 (alpha subunit) |
| chr14_+_54082691 | 0.50 |
ENSMUST00000103674.6
|
Trav19
|
T cell receptor alpha variable 19 |
| chr4_-_149211145 | 0.49 |
ENSMUST00000030815.3
|
Cort
|
cortistatin |
| chr1_+_87254719 | 0.49 |
ENSMUST00000027475.15
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr14_+_53048391 | 0.47 |
ENSMUST00000103646.5
|
Trav10d
|
T cell receptor alpha variable 10D |
| chr4_-_133728168 | 0.46 |
ENSMUST00000105887.8
ENSMUST00000012262.12 ENSMUST00000144668.8 ENSMUST00000105889.4 |
Dhdds
|
dehydrodolichyl diphosphate synthase |
| chr6_+_34686543 | 0.46 |
ENSMUST00000031775.13
|
Cald1
|
caldesmon 1 |
| chr5_-_27706360 | 0.46 |
ENSMUST00000155721.2
ENSMUST00000053257.10 |
Speer4b
|
spermatogenesis associated glutamate (E)-rich protein 4B |
| chr7_+_13132040 | 0.46 |
ENSMUST00000005791.14
|
Cabp5
|
calcium binding protein 5 |
| chr5_-_26193648 | 0.46 |
ENSMUST00000191203.7
|
Gm21698
|
predicted gene, 21698 |
| chr5_-_26294289 | 0.46 |
ENSMUST00000094946.5
|
Gm10471
|
predicted gene 10471 |
| chr13_-_21765822 | 0.45 |
ENSMUST00000206526.3
ENSMUST00000213912.2 |
Olfr1364
|
olfactory receptor 1364 |
| chr17_+_38231439 | 0.45 |
ENSMUST00000216440.2
|
Olfr128
|
olfactory receptor 128 |
| chr12_+_65272495 | 0.45 |
ENSMUST00000221980.2
|
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr9_-_121324705 | 0.44 |
ENSMUST00000216138.2
|
Cck
|
cholecystokinin |
| chr14_+_115329676 | 0.44 |
ENSMUST00000176912.8
ENSMUST00000175665.8 |
Gpc5
|
glypican 5 |
| chr9_+_3036877 | 0.44 |
ENSMUST00000155807.3
|
Gm10715
|
predicted gene 10715 |
| chr7_-_43906629 | 0.43 |
ENSMUST00000012921.9
|
Acp4
|
acid phosphatase 4 |
| chr14_-_72840373 | 0.43 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr6_+_40546421 | 0.42 |
ENSMUST00000216942.2
|
Olfr460
|
olfactory receptor 460 |
| chr4_-_114991478 | 0.42 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr8_-_106306477 | 0.41 |
ENSMUST00000194654.2
|
Agrp
|
agouti related neuropeptide |
| chr7_+_44592628 | 0.41 |
ENSMUST00000207719.2
ENSMUST00000120929.9 |
Tsks
|
testis-specific serine kinase substrate |
| chr17_+_38211328 | 0.41 |
ENSMUST00000217223.2
|
Olfr127
|
olfactory receptor 127 |
| chr7_-_21750929 | 0.39 |
ENSMUST00000178209.2
|
Gm6176
|
predicted gene 6176 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hdx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 28.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 3.9 | 35.0 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 3.3 | 9.9 | GO:0097037 | heme export(GO:0097037) |
| 2.8 | 16.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.8 | 7.1 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.4 | 12.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 1.1 | 20.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 1.0 | 3.8 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.8 | 10.0 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.7 | 4.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.6 | 5.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.6 | 2.8 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.5 | 1.5 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.5 | 1.5 | GO:1904154 | trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.5 | 2.3 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.4 | 1.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.4 | 7.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 1.8 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 1.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 2.3 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.3 | 2.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 1.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.3 | 1.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 2.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 1.7 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 2.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 3.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.0 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 2.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 3.5 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 1.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 2.9 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 0.9 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.5 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 | 1.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 | 3.8 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 2.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.7 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 1.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.1 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 2.0 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 4.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.2 | GO:0070318 | enucleate erythrocyte development(GO:0048822) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 0.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 9.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 0.5 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.1 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 2.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 3.1 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.5 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 1.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 1.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 7.7 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 5.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 1.4 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.7 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.6 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 4.8 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.7 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.0 | 1.5 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 4.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.9 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 1.3 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.9 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.6 | GO:0048806 | genitalia development(GO:0048806) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 3.2 | 28.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.6 | 4.7 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.5 | 2.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 3.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.3 | 11.1 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.2 | 2.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 2.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 7.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 2.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 2.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 7.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 9.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 22.9 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 7.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 32.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 3.3 | 16.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.8 | 12.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.5 | 12.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.4 | 7.1 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 1.3 | 3.8 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 1.0 | 20.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.8 | 2.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.6 | 1.8 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.6 | 23.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.5 | 3.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 1.8 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 2.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 7.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 1.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 3.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 0.6 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.2 | 5.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 0.7 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 18.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 1.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 4.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 1.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 2.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 2.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 4.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.4 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 16.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 17.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 9.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 6.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 32.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.4 | 20.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 1.3 | 40.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.9 | 10.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.6 | 16.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.5 | 7.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 7.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 2.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 4.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 9.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 2.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 2.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.5 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |