Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
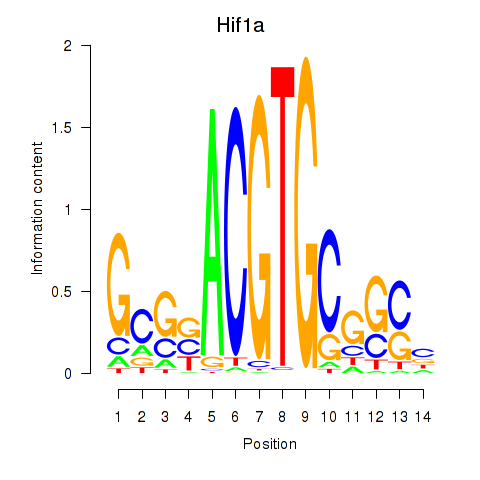
Results for Hif1a
Z-value: 1.34
Transcription factors associated with Hif1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hif1a
|
ENSMUSG00000021109.14 | hypoxia inducible factor 1, alpha subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hif1a | mm39_v1_chr12_+_73948143_73948166 | -0.46 | 4.4e-03 | Click! |
Activity profile of Hif1a motif
Sorted Z-values of Hif1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_120839879 | 7.34 |
ENSMUST00000154187.8
ENSMUST00000100130.10 ENSMUST00000129473.8 ENSMUST00000168579.8 |
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chrX_-_92875712 | 6.04 |
ENSMUST00000045748.7
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr5_+_129097133 | 5.81 |
ENSMUST00000031383.14
ENSMUST00000111343.2 |
Ran
|
RAN, member RAS oncogene family |
| chr17_+_28988354 | 5.61 |
ENSMUST00000233109.2
ENSMUST00000004986.14 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr17_+_28988271 | 5.19 |
ENSMUST00000233984.2
ENSMUST00000233460.2 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr8_-_46452896 | 4.98 |
ENSMUST00000053558.10
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr6_-_70769135 | 4.65 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr8_-_123278054 | 4.62 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr11_+_53991750 | 4.46 |
ENSMUST00000093107.12
ENSMUST00000019050.12 ENSMUST00000174616.8 ENSMUST00000129499.8 ENSMUST00000126840.8 |
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr8_-_122634418 | 4.29 |
ENSMUST00000045557.10
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chr11_+_72851989 | 4.08 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr11_+_61575245 | 3.62 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G |
| chr6_+_29272625 | 3.40 |
ENSMUST00000054445.9
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr4_-_43045685 | 3.38 |
ENSMUST00000107956.8
ENSMUST00000107957.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr5_+_33815466 | 3.34 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr2_+_155359868 | 3.19 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr4_-_43046196 | 3.15 |
ENSMUST00000036462.12
|
Fam214b
|
family with sequence similarity 214, member B |
| chr10_-_59787646 | 3.09 |
ENSMUST00000020308.5
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chr8_+_84724130 | 2.92 |
ENSMUST00000095228.5
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr6_+_29272463 | 2.88 |
ENSMUST00000115289.2
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr12_+_104998895 | 2.87 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr10_-_77845571 | 2.80 |
ENSMUST00000020522.9
|
Pfkl
|
phosphofructokinase, liver, B-type |
| chr2_+_127178072 | 2.79 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chr7_+_28240262 | 2.77 |
ENSMUST00000119180.4
|
Sycn
|
syncollin |
| chr2_+_155360015 | 2.68 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr16_-_18448614 | 2.62 |
ENSMUST00000231956.2
ENSMUST00000096987.7 |
Septin5
|
septin 5 |
| chr3_-_107993906 | 2.61 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chrX_+_134894573 | 2.59 |
ENSMUST00000058119.9
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr17_-_26314461 | 2.59 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr17_-_26314438 | 2.46 |
ENSMUST00000236547.2
|
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr11_+_102080446 | 2.30 |
ENSMUST00000070334.10
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr2_+_30697838 | 2.21 |
ENSMUST00000041830.10
ENSMUST00000152374.8 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr9_+_107464841 | 2.06 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr6_+_125108829 | 2.04 |
ENSMUST00000044200.11
ENSMUST00000204185.2 |
Nop2
|
NOP2 nucleolar protein |
| chr11_+_102080489 | 2.03 |
ENSMUST00000078975.8
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr2_-_14060774 | 1.96 |
ENSMUST00000114753.8
ENSMUST00000091429.12 |
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr11_+_68989763 | 1.94 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr1_+_63216281 | 1.83 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr11_-_120239339 | 1.80 |
ENSMUST00000071555.13
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr1_+_172327812 | 1.79 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr9_+_106080307 | 1.76 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr7_+_46495521 | 1.72 |
ENSMUST00000133062.2
|
Ldha
|
lactate dehydrogenase A |
| chr2_-_14060840 | 1.71 |
ENSMUST00000074854.9
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr11_-_120239301 | 1.70 |
ENSMUST00000062147.14
ENSMUST00000128055.2 |
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr9_-_57672375 | 1.68 |
ENSMUST00000215233.2
|
Clk3
|
CDC-like kinase 3 |
| chr11_-_88608920 | 1.67 |
ENSMUST00000092794.12
|
Msi2
|
musashi RNA-binding protein 2 |
| chr1_+_63215976 | 1.66 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr4_-_43523595 | 1.66 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr4_-_129494435 | 1.64 |
ENSMUST00000102593.11
|
Eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr15_-_75781138 | 1.63 |
ENSMUST00000145764.2
ENSMUST00000116440.9 ENSMUST00000151066.8 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr16_-_18448454 | 1.62 |
ENSMUST00000231622.2
|
Septin5
|
septin 5 |
| chr16_+_49675969 | 1.60 |
ENSMUST00000229101.2
ENSMUST00000230836.2 ENSMUST00000229206.2 ENSMUST00000084838.14 ENSMUST00000230281.2 |
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr4_-_129494378 | 1.59 |
ENSMUST00000135055.8
|
Eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr11_-_120238917 | 1.57 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr2_-_31973795 | 1.56 |
ENSMUST00000056406.7
|
Fam78a
|
family with sequence similarity 78, member A |
| chr7_+_3706992 | 1.53 |
ENSMUST00000006496.15
ENSMUST00000108623.8 ENSMUST00000139818.2 ENSMUST00000108625.8 |
Rps9
|
ribosomal protein S9 |
| chr8_+_107620251 | 1.52 |
ENSMUST00000212272.2
ENSMUST00000047629.7 |
Utp4
|
UTP4 small subunit processome component |
| chr15_-_75781387 | 1.52 |
ENSMUST00000123712.8
ENSMUST00000141475.2 ENSMUST00000144614.8 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr11_-_116001037 | 1.52 |
ENSMUST00000106441.8
ENSMUST00000021120.6 |
Trim47
|
tripartite motif-containing 47 |
| chr8_-_107620210 | 1.51 |
ENSMUST00000177068.8
ENSMUST00000176515.2 ENSMUST00000169312.2 |
Chtf8
Derpc
|
CTF8, chromosome transmission fidelity factor 8 DERPC proline and glycine rich nuclear protein |
| chr11_+_117006020 | 1.51 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chr15_-_75781168 | 1.50 |
ENSMUST00000089680.10
ENSMUST00000141268.8 ENSMUST00000023235.13 ENSMUST00000109972.9 ENSMUST00000089681.12 ENSMUST00000109975.10 ENSMUST00000154584.9 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr7_+_46495256 | 1.48 |
ENSMUST00000048209.16
ENSMUST00000210815.2 ENSMUST00000125862.8 ENSMUST00000210968.2 ENSMUST00000092621.12 ENSMUST00000210467.2 |
Ldha
|
lactate dehydrogenase A |
| chr11_+_53992054 | 1.46 |
ENSMUST00000135653.8
|
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr11_+_78069477 | 1.44 |
ENSMUST00000092880.14
ENSMUST00000127587.8 |
Tlcd1
|
TLC domain containing 1 |
| chr13_+_91071077 | 1.44 |
ENSMUST00000051955.9
|
Rps23
|
ribosomal protein S23 |
| chr11_+_117005958 | 1.42 |
ENSMUST00000021177.15
|
Sec14l1
|
SEC14-like lipid binding 1 |
| chr4_-_62438122 | 1.39 |
ENSMUST00000107444.8
ENSMUST00000030090.4 |
Alad
|
aminolevulinate, delta-, dehydratase |
| chr16_+_18695787 | 1.38 |
ENSMUST00000120532.9
ENSMUST00000004222.14 |
Hira
|
histone cell cycle regulator |
| chr19_+_6952580 | 1.36 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr7_+_105385772 | 1.35 |
ENSMUST00000033182.10
|
Ilk
|
integrin linked kinase |
| chr7_+_105385864 | 1.33 |
ENSMUST00000163389.9
ENSMUST00000136687.9 |
Ilk
|
integrin linked kinase |
| chr19_+_6952319 | 1.31 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr11_+_121150798 | 1.26 |
ENSMUST00000106113.2
|
Foxk2
|
forkhead box K2 |
| chr7_-_44635740 | 1.26 |
ENSMUST00000209056.3
ENSMUST00000209124.2 ENSMUST00000208312.2 ENSMUST00000207659.2 ENSMUST00000045325.14 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr6_+_85428464 | 1.24 |
ENSMUST00000032078.9
|
Cct7
|
chaperonin containing Tcp1, subunit 7 (eta) |
| chr10_+_80591030 | 1.21 |
ENSMUST00000105336.9
|
Dot1l
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae) |
| chr7_-_44635813 | 1.21 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr4_-_139079842 | 1.19 |
ENSMUST00000102503.10
|
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr12_-_40087393 | 1.19 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr17_-_27352593 | 1.18 |
ENSMUST00000118613.8
|
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr17_+_73144531 | 1.16 |
ENSMUST00000233886.2
|
Ypel5
|
yippee like 5 |
| chr10_+_80100812 | 1.14 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr9_-_57168777 | 1.13 |
ENSMUST00000217657.2
|
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr4_-_139079609 | 1.13 |
ENSMUST00000030513.13
ENSMUST00000155257.8 |
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr14_+_121148625 | 1.09 |
ENSMUST00000032898.9
|
Ipo5
|
importin 5 |
| chr11_-_72302520 | 1.08 |
ENSMUST00000108500.8
ENSMUST00000050226.7 |
Smtnl2
|
smoothelin-like 2 |
| chr4_+_152410291 | 1.06 |
ENSMUST00000103191.11
ENSMUST00000139685.8 ENSMUST00000188151.2 |
Rpl22
|
ribosomal protein L22 |
| chr10_+_80100868 | 1.01 |
ENSMUST00000092305.6
|
Dazap1
|
DAZ associated protein 1 |
| chr1_+_78286946 | 1.01 |
ENSMUST00000036172.10
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr19_+_47167259 | 1.00 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr7_-_126101555 | 1.00 |
ENSMUST00000167759.8
|
Atxn2l
|
ataxin 2-like |
| chr4_+_150321272 | 0.99 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr8_-_107620147 | 0.99 |
ENSMUST00000176090.2
|
Chtf8
|
CTF8, chromosome transmission fidelity factor 8 |
| chr14_+_33041660 | 0.99 |
ENSMUST00000111955.2
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr11_+_3913970 | 0.98 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr15_-_102112159 | 0.98 |
ENSMUST00000229252.2
ENSMUST00000229770.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr10_+_122514669 | 0.95 |
ENSMUST00000161487.8
ENSMUST00000067918.12 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr7_-_126101245 | 0.94 |
ENSMUST00000179818.3
|
Atxn2l
|
ataxin 2-like |
| chr1_+_89507952 | 0.93 |
ENSMUST00000074945.8
|
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr15_-_36609208 | 0.93 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr11_-_120472379 | 0.92 |
ENSMUST00000106197.10
|
Arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr7_-_126101484 | 0.89 |
ENSMUST00000166682.9
|
Atxn2l
|
ataxin 2-like |
| chr11_-_120472022 | 0.87 |
ENSMUST00000067936.6
|
Arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr2_+_27405169 | 0.84 |
ENSMUST00000113952.10
|
Wdr5
|
WD repeat domain 5 |
| chr7_+_19311212 | 0.84 |
ENSMUST00000108453.2
|
Zfp296
|
zinc finger protein 296 |
| chr2_+_162773440 | 0.82 |
ENSMUST00000130411.7
ENSMUST00000126163.3 |
Srsf6
|
serine and arginine-rich splicing factor 6 |
| chr11_-_88609048 | 0.80 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr17_-_27352876 | 0.80 |
ENSMUST00000119227.3
ENSMUST00000025045.15 |
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr6_-_85428207 | 0.77 |
ENSMUST00000113770.2
ENSMUST00000032080.9 |
Pradc1
|
protease-associated domain containing 1 |
| chr11_+_78069528 | 0.77 |
ENSMUST00000108338.2
|
Tlcd1
|
TLC domain containing 1 |
| chr5_-_148988413 | 0.77 |
ENSMUST00000093196.11
|
Hmgb1
|
high mobility group box 1 |
| chr10_-_80223475 | 0.72 |
ENSMUST00000105350.3
|
Mex3d
|
mex3 RNA binding family member D |
| chr11_-_88608958 | 0.72 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr9_-_110483210 | 0.72 |
ENSMUST00000196488.5
ENSMUST00000133191.8 ENSMUST00000167320.8 |
Nbeal2
|
neurobeachin-like 2 |
| chr10_-_84938350 | 0.71 |
ENSMUST00000059383.8
ENSMUST00000216889.2 |
Fhl4
|
four and a half LIM domains 4 |
| chr10_-_81436671 | 0.71 |
ENSMUST00000151858.8
ENSMUST00000142948.8 ENSMUST00000072020.9 |
Tle6
|
transducin-like enhancer of split 6 |
| chr15_-_36609812 | 0.70 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr15_+_85942928 | 0.70 |
ENSMUST00000138134.8
|
Gramd4
|
GRAM domain containing 4 |
| chr4_+_154255187 | 0.69 |
ENSMUST00000030897.15
|
Megf6
|
multiple EGF-like-domains 6 |
| chr17_-_46991709 | 0.69 |
ENSMUST00000233524.2
ENSMUST00000233733.2 ENSMUST00000071841.7 ENSMUST00000165007.9 |
Klhdc3
|
kelch domain containing 3 |
| chr4_+_150321142 | 0.68 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr9_-_96246460 | 0.67 |
ENSMUST00000034983.7
|
Atp1b3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr5_-_124387812 | 0.66 |
ENSMUST00000162812.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr8_+_34222058 | 0.65 |
ENSMUST00000167264.8
ENSMUST00000187392.7 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr7_+_113113061 | 0.65 |
ENSMUST00000129087.8
ENSMUST00000067929.15 ENSMUST00000164745.8 ENSMUST00000136158.8 |
Far1
|
fatty acyl CoA reductase 1 |
| chr11_+_3282424 | 0.61 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr17_+_27276262 | 0.60 |
ENSMUST00000049308.9
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr4_-_133695264 | 0.58 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr8_+_34221861 | 0.58 |
ENSMUST00000170705.8
|
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr8_+_34222266 | 0.58 |
ENSMUST00000190675.2
ENSMUST00000171010.8 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr1_-_162376053 | 0.56 |
ENSMUST00000028017.16
|
Eef1aknmt
|
EEF1A lysine and N-terminal methyltransferase |
| chr13_-_117161921 | 0.55 |
ENSMUST00000223949.2
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr19_+_47167554 | 0.54 |
ENSMUST00000235290.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr15_-_78088423 | 0.53 |
ENSMUST00000005860.16
|
Pvalb
|
parvalbumin |
| chr10_+_80062468 | 0.52 |
ENSMUST00000130260.2
|
Pwwp3a
|
PWWP domain containing 3A, DNA repair factor |
| chr13_-_117162041 | 0.50 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr11_+_87017878 | 0.50 |
ENSMUST00000041282.13
|
Trim37
|
tripartite motif-containing 37 |
| chr16_+_20317544 | 0.48 |
ENSMUST00000003320.14
|
Eif2b5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon |
| chr2_+_127967951 | 0.47 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr5_-_135280063 | 0.47 |
ENSMUST00000062572.3
|
Fzd9
|
frizzled class receptor 9 |
| chr7_-_138511221 | 0.46 |
ENSMUST00000130500.8
ENSMUST00000106112.2 |
Bnip3
|
BCL2/adenovirus E1B interacting protein 3 |
| chr16_-_20060053 | 0.44 |
ENSMUST00000040880.9
|
Map6d1
|
MAP6 domain containing 1 |
| chr11_+_68582731 | 0.44 |
ENSMUST00000102611.10
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr3_+_122039274 | 0.43 |
ENSMUST00000178826.8
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr4_+_11191354 | 0.43 |
ENSMUST00000170901.8
|
Ccne2
|
cyclin E2 |
| chr5_+_14075281 | 0.41 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr14_-_30741012 | 0.41 |
ENSMUST00000037739.8
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr11_-_87249837 | 0.40 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr2_+_91357100 | 0.36 |
ENSMUST00000111338.10
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr11_+_68582923 | 0.35 |
ENSMUST00000018887.15
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr18_+_35686424 | 0.35 |
ENSMUST00000235679.2
ENSMUST00000235176.2 ENSMUST00000235801.2 ENSMUST00000237592.2 ENSMUST00000237230.2 ENSMUST00000237589.2 |
Snhg4
Snhg4
|
small nucleolar RNA host gene 4 small nucleolar RNA host gene 4 |
| chr2_+_151544086 | 0.35 |
ENSMUST00000109872.2
|
Tmem74b
|
transmembrane protein 74B |
| chr1_+_39232812 | 0.34 |
ENSMUST00000173050.8
|
Npas2
|
neuronal PAS domain protein 2 |
| chr7_+_28392456 | 0.33 |
ENSMUST00000151227.2
ENSMUST00000108281.8 |
Fbxo27
|
F-box protein 27 |
| chr10_+_128073900 | 0.33 |
ENSMUST00000105245.3
|
Timeless
|
timeless circadian clock 1 |
| chr2_-_105229653 | 0.32 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr15_+_80861966 | 0.31 |
ENSMUST00000139517.9
ENSMUST00000137255.3 ENSMUST00000137004.2 |
Sgsm3
|
small G protein signaling modulator 3 |
| chr1_+_55127110 | 0.31 |
ENSMUST00000075242.7
|
Hspe1
|
heat shock protein 1 (chaperonin 10) |
| chr5_-_23988551 | 0.31 |
ENSMUST00000148618.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr11_+_87018079 | 0.30 |
ENSMUST00000139532.2
|
Trim37
|
tripartite motif-containing 37 |
| chr12_+_51394812 | 0.30 |
ENSMUST00000054308.13
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chrX_+_10583629 | 0.29 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr3_+_122039206 | 0.29 |
ENSMUST00000029769.14
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr16_+_18066825 | 0.28 |
ENSMUST00000100099.10
|
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr11_-_95956176 | 0.27 |
ENSMUST00000100528.5
|
Ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr9_+_108225026 | 0.25 |
ENSMUST00000035237.12
ENSMUST00000194959.6 |
Usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr7_+_113113037 | 0.24 |
ENSMUST00000033018.15
|
Far1
|
fatty acyl CoA reductase 1 |
| chr5_-_123662175 | 0.23 |
ENSMUST00000200247.5
ENSMUST00000111586.8 ENSMUST00000031385.7 ENSMUST00000145152.8 ENSMUST00000111587.10 |
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr16_+_23807232 | 0.23 |
ENSMUST00000210705.2
|
Gm37419
|
predicted gene, 37419 |
| chr16_+_18066730 | 0.23 |
ENSMUST00000115640.8
ENSMUST00000140206.8 |
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr19_+_47167444 | 0.22 |
ENSMUST00000235326.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr5_-_23988696 | 0.21 |
ENSMUST00000119946.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr13_+_38529062 | 0.21 |
ENSMUST00000171970.3
|
Bmp6
|
bone morphogenetic protein 6 |
| chr2_+_164327988 | 0.21 |
ENSMUST00000109350.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr10_+_17598961 | 0.21 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chrX_-_72868544 | 0.21 |
ENSMUST00000002080.12
ENSMUST00000114438.3 |
Pdzd4
|
PDZ domain containing 4 |
| chr5_-_135092904 | 0.20 |
ENSMUST00000111205.8
ENSMUST00000141309.2 |
Bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr4_+_129494463 | 0.19 |
ENSMUST00000102591.10
ENSMUST00000181579.8 ENSMUST00000173758.8 |
Tmem234
|
transmembrane protein 234 |
| chr18_+_52598748 | 0.19 |
ENSMUST00000025406.9
|
Srfbp1
|
serum response factor binding protein 1 |
| chr16_-_18066591 | 0.19 |
ENSMUST00000115645.10
|
Ranbp1
|
RAN binding protein 1 |
| chr15_+_80171435 | 0.19 |
ENSMUST00000160424.8
|
Cacna1i
|
calcium channel, voltage-dependent, alpha 1I subunit |
| chr14_-_70680659 | 0.19 |
ENSMUST00000180358.3
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr12_+_117497478 | 0.19 |
ENSMUST00000220781.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr14_-_70680882 | 0.18 |
ENSMUST00000000793.13
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr8_-_88199231 | 0.16 |
ENSMUST00000034076.16
|
Cbln1
|
cerebellin 1 precursor protein |
| chr17_-_45884179 | 0.16 |
ENSMUST00000165127.8
ENSMUST00000166469.8 ENSMUST00000024739.14 |
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr1_+_185187000 | 0.16 |
ENSMUST00000061093.7
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr17_-_23896394 | 0.16 |
ENSMUST00000233428.2
ENSMUST00000233814.2 ENSMUST00000167059.9 ENSMUST00000024698.10 |
Tnfrsf12a
|
tumor necrosis factor receptor superfamily, member 12a |
| chr5_+_100187844 | 0.16 |
ENSMUST00000169390.8
ENSMUST00000031268.8 |
Enoph1
|
enolase-phosphatase 1 |
| chr7_-_136915602 | 0.15 |
ENSMUST00000210774.2
|
Ebf3
|
early B cell factor 3 |
| chr13_-_100969823 | 0.14 |
ENSMUST00000225922.2
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr15_+_80862074 | 0.14 |
ENSMUST00000229727.2
|
Sgsm3
|
small G protein signaling modulator 3 |
| chr8_+_65399831 | 0.14 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr4_+_150321659 | 0.14 |
ENSMUST00000133839.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr12_+_49431957 | 0.14 |
ENSMUST00000110746.2
|
Gm43517
|
predicted gene 43517 |
| chr13_-_72111832 | 0.13 |
ENSMUST00000077337.9
|
Irx1
|
Iroquois homeobox 1 |
| chr1_-_153425791 | 0.13 |
ENSMUST00000041874.9
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr6_-_38614155 | 0.12 |
ENSMUST00000096030.7
ENSMUST00000201345.2 |
Klrg2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr19_+_44994905 | 0.12 |
ENSMUST00000026227.3
|
Twnk
|
twinkle mtDNA helicase |
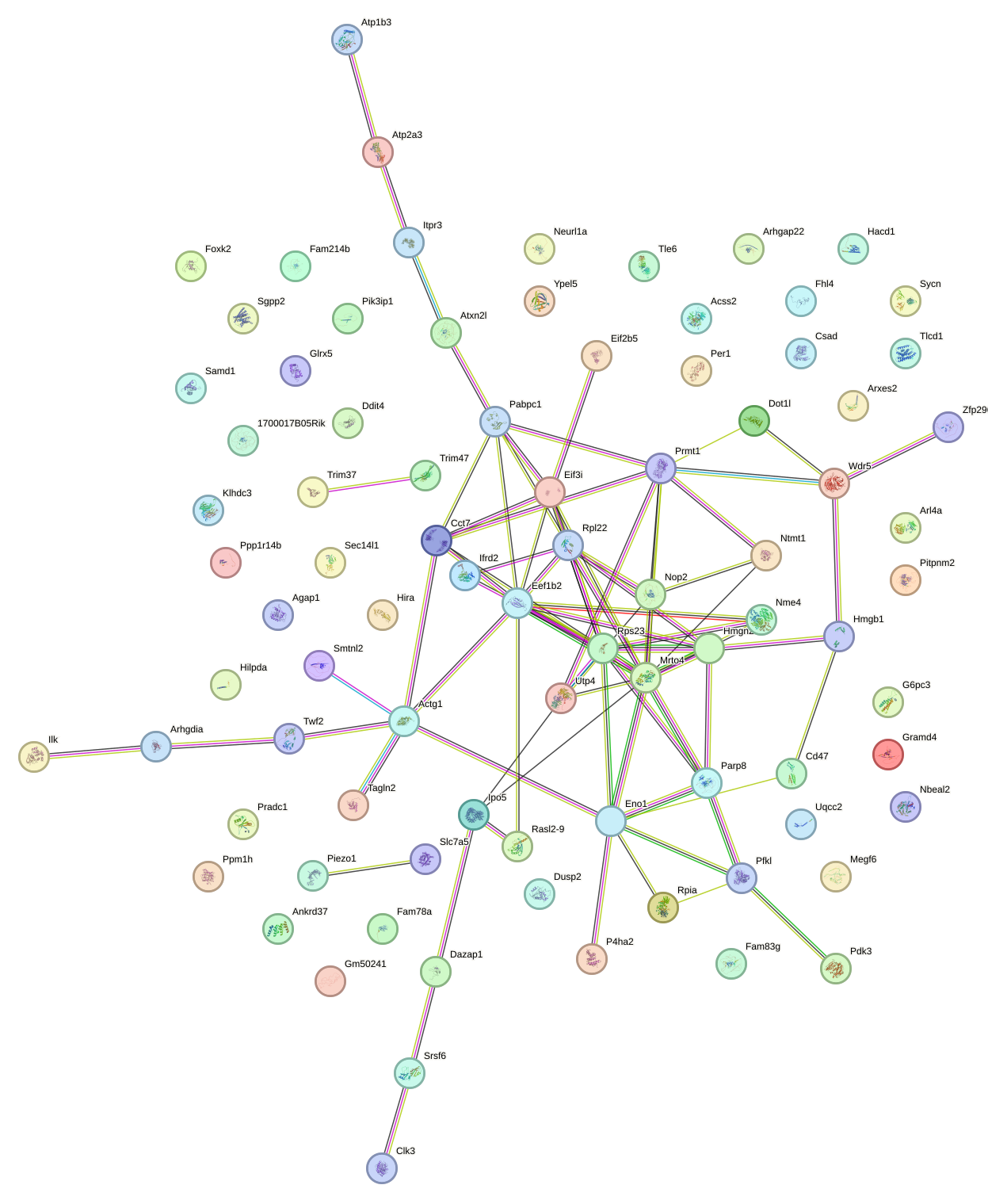
Network of associatons between targets according to the STRING database.
First level regulatory network of Hif1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 2.0 | 5.9 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 1.5 | 6.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.2 | 4.7 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.9 | 4.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.7 | 5.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.6 | 7.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.6 | 5.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.6 | 2.2 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.5 | 2.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.4 | 4.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 2.9 | GO:0015871 | choline transport(GO:0015871) |
| 0.4 | 5.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 1.9 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.3 | 3.2 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 | 2.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 2.5 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 1.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 2.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 0.8 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.2 | 2.8 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 1.6 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 2.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 1.0 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.2 | 3.1 | GO:0051198 | negative regulation of nucleotide catabolic process(GO:0030812) negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.2 | 1.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 2.0 | GO:0070131 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 0.5 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 2.8 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 8.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.9 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 3.7 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.4 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 1.6 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 3.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.1 | 1.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.7 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 1.8 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.7 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.6 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.5 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.5 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.1 | 1.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 2.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 4.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.2 | GO:1904867 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 9.1 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 0.7 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 1.4 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.1 | 2.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.9 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.8 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 1.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 2.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.2 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 4.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 2.2 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 1.0 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 2.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 1.4 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.2 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 1.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 1.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 1.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 3.2 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 2.6 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 4.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 2.3 | GO:0045444 | fat cell differentiation(GO:0045444) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 1.2 | 5.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.2 | 8.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 1.8 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.6 | 2.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.5 | 1.5 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 3.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 2.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 0.7 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 2.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 7.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 4.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 7.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 3.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 1.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 9.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.2 | 3.7 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 1.2 | 5.9 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.1 | 4.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.0 | 5.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.8 | 5.8 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.8 | 7.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.7 | 2.0 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.6 | 2.5 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.6 | 2.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.5 | 5.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.5 | 4.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 1.8 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.4 | 4.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 1.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.4 | 9.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 8.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 3.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 2.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 0.8 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.3 | 1.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.7 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.2 | 2.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 2.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 4.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 0.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 4.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 1.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.0 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 1.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 2.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 2.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 4.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 3.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 4.1 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 2.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 6.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 1.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 5.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 5.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 9.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 8.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 3.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 14.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.5 | 5.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.5 | 9.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 5.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 4.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 5.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 5.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 6.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 4.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 4.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 9.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 2.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |