Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
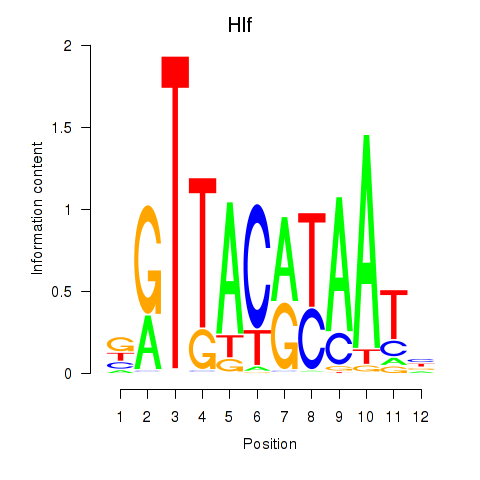
Results for Hlf
Z-value: 1.11
Transcription factors associated with Hlf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hlf
|
ENSMUSG00000003949.17 | hepatic leukemia factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hlf | mm39_v1_chr11_-_90281721_90281753 | 0.45 | 5.5e-03 | Click! |
Activity profile of Hlf motif
Sorted Z-values of Hlf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_22738987 | 14.38 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr16_+_22769822 | 11.41 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr6_+_121815473 | 11.03 |
ENSMUST00000032228.9
|
Mug1
|
murinoglobulin 1 |
| chr10_+_23727325 | 9.63 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr16_+_22769844 | 9.58 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr7_+_26534730 | 9.13 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr14_+_66208498 | 8.92 |
ENSMUST00000128539.8
|
Clu
|
clusterin |
| chr7_+_25872836 | 8.87 |
ENSMUST00000082214.5
|
Cyp2b9
|
cytochrome P450, family 2, subfamily b, polypeptide 9 |
| chr12_-_81014755 | 8.55 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr16_+_22739191 | 8.50 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr16_+_22739028 | 7.88 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chr14_+_66208613 | 7.85 |
ENSMUST00000144619.2
|
Clu
|
clusterin |
| chr11_+_115353290 | 7.33 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr7_-_46392403 | 7.23 |
ENSMUST00000128088.4
|
Saa1
|
serum amyloid A 1 |
| chr5_-_87074380 | 6.18 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr7_+_26006594 | 6.14 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr7_-_26638802 | 5.90 |
ENSMUST00000170227.3
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22 |
| chr9_-_71070506 | 5.50 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr12_-_81014849 | 5.47 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr14_+_40827317 | 5.46 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr7_+_25760922 | 5.44 |
ENSMUST00000005669.9
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr9_-_78254422 | 5.40 |
ENSMUST00000034902.12
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr1_-_140111138 | 4.89 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 4.75 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr15_-_3612078 | 4.73 |
ENSMUST00000161770.2
|
Ghr
|
growth hormone receptor |
| chr12_-_104831266 | 4.70 |
ENSMUST00000109937.9
|
Clmn
|
calmin |
| chr8_-_118398264 | 4.69 |
ENSMUST00000037955.14
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr4_-_49408040 | 4.65 |
ENSMUST00000081541.9
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr15_-_76191301 | 4.63 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr7_+_46401214 | 4.34 |
ENSMUST00000210769.2
ENSMUST00000210272.2 ENSMUST00000075982.4 |
Saa2
|
serum amyloid A 2 |
| chr12_-_104831335 | 4.13 |
ENSMUST00000109936.3
|
Clmn
|
calmin |
| chr3_-_113368407 | 4.09 |
ENSMUST00000106540.8
|
Amy1
|
amylase 1, salivary |
| chr9_-_78254443 | 4.07 |
ENSMUST00000129247.2
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr10_-_95678748 | 4.07 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr7_-_30643444 | 4.05 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr13_+_25127127 | 3.69 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr10_-_125225298 | 3.65 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr5_-_87240405 | 3.40 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr16_+_37400590 | 3.38 |
ENSMUST00000159787.8
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr16_+_37400500 | 3.35 |
ENSMUST00000160847.2
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr10_-_95678786 | 3.27 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr5_-_92475927 | 3.24 |
ENSMUST00000113093.5
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr15_+_99291491 | 3.14 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr15_+_99291455 | 3.11 |
ENSMUST00000162624.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr19_+_58717319 | 3.01 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr19_+_38995463 | 2.99 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr8_-_93956143 | 2.92 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr19_-_7779943 | 2.87 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr14_+_70815250 | 2.82 |
ENSMUST00000228554.2
|
Nudt18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr9_-_107546195 | 2.81 |
ENSMUST00000192990.6
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr10_-_78300802 | 2.74 |
ENSMUST00000041616.15
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr5_+_135038267 | 2.72 |
ENSMUST00000201890.2
ENSMUST00000154469.8 |
Abhd11
|
abhydrolase domain containing 11 |
| chr8_+_86219191 | 2.72 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr2_+_155118217 | 2.71 |
ENSMUST00000029128.4
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr8_+_46924074 | 2.70 |
ENSMUST00000034046.13
ENSMUST00000211644.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr9_-_107546166 | 2.68 |
ENSMUST00000177567.8
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr17_+_79922329 | 2.64 |
ENSMUST00000040368.3
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr16_-_22847808 | 2.58 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr6_+_113460258 | 2.55 |
ENSMUST00000032422.6
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr16_-_22847760 | 2.53 |
ENSMUST00000039338.13
|
Kng2
|
kininogen 2 |
| chr9_+_107957621 | 2.52 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr16_-_22847829 | 2.47 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr9_+_107957640 | 2.46 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr19_-_7780025 | 2.46 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr5_-_5564730 | 2.45 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr5_+_135038006 | 2.42 |
ENSMUST00000111216.8
ENSMUST00000046999.12 |
Abhd11
|
abhydrolase domain containing 11 |
| chr18_-_35631914 | 2.37 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr4_+_47208004 | 2.36 |
ENSMUST00000082303.13
ENSMUST00000102917.11 |
Col15a1
|
collagen, type XV, alpha 1 |
| chr14_+_79086492 | 2.29 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr16_-_38253507 | 2.27 |
ENSMUST00000002926.8
|
Pla1a
|
phospholipase A1 member A |
| chr5_-_5564873 | 2.27 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr7_+_67297152 | 2.22 |
ENSMUST00000032774.16
ENSMUST00000107471.8 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr10_-_31321793 | 2.17 |
ENSMUST00000213639.2
ENSMUST00000215515.2 ENSMUST00000214644.2 ENSMUST00000213528.2 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr10_+_21253190 | 2.16 |
ENSMUST00000042699.14
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr12_+_37291632 | 2.12 |
ENSMUST00000049874.14
|
Agmo
|
alkylglycerol monooxygenase |
| chr16_+_3702604 | 2.11 |
ENSMUST00000115860.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr7_-_126873219 | 2.09 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr1_-_71692320 | 2.08 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr2_+_118692435 | 2.07 |
ENSMUST00000028807.6
|
Ivd
|
isovaleryl coenzyme A dehydrogenase |
| chr16_+_3702523 | 2.06 |
ENSMUST00000176625.8
ENSMUST00000186375.8 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr1_-_171173198 | 2.05 |
ENSMUST00000111295.8
ENSMUST00000148339.2 ENSMUST00000111289.8 |
Nit1
|
nitrilase 1 |
| chr12_+_37291728 | 2.00 |
ENSMUST00000160768.8
|
Agmo
|
alkylglycerol monooxygenase |
| chr14_-_59835285 | 1.99 |
ENSMUST00000022555.11
ENSMUST00000225839.2 ENSMUST00000056997.15 ENSMUST00000171683.3 ENSMUST00000167100.9 |
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr10_+_78410180 | 1.98 |
ENSMUST00000218061.2
ENSMUST00000218787.2 ENSMUST00000105384.5 ENSMUST00000218875.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr15_-_58953838 | 1.95 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr1_-_171173168 | 1.94 |
ENSMUST00000156856.8
ENSMUST00000111296.8 |
Nit1
|
nitrilase 1 |
| chr16_-_87229485 | 1.90 |
ENSMUST00000039449.9
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr16_-_22848153 | 1.90 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr16_+_3702493 | 1.89 |
ENSMUST00000176233.2
|
Gm20695
|
predicted gene 20695 |
| chr3_+_59939175 | 1.86 |
ENSMUST00000029325.5
|
Aadac
|
arylacetamide deacetylase |
| chr5_-_147831610 | 1.82 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr6_-_115569504 | 1.81 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr10_-_115197775 | 1.80 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr9_+_78197205 | 1.79 |
ENSMUST00000119823.8
ENSMUST00000121273.2 |
Gsta5
|
glutathione S-transferase alpha 5 |
| chr3_-_129126362 | 1.77 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr11_-_69576363 | 1.72 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr16_-_4608084 | 1.71 |
ENSMUST00000118703.8
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr19_+_34268053 | 1.69 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr5_-_31265562 | 1.66 |
ENSMUST00000201396.2
ENSMUST00000202740.4 |
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chrX_+_93278203 | 1.62 |
ENSMUST00000153900.8
|
Klhl15
|
kelch-like 15 |
| chr16_-_4607751 | 1.62 |
ENSMUST00000117713.8
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr13_+_114954741 | 1.59 |
ENSMUST00000166104.9
ENSMUST00000166176.9 ENSMUST00000184335.8 ENSMUST00000184245.8 ENSMUST00000015680.11 ENSMUST00000184214.8 ENSMUST00000165022.9 ENSMUST00000164737.8 ENSMUST00000184781.8 ENSMUST00000183407.2 ENSMUST00000184672.8 |
Mocs2
|
molybdenum cofactor synthesis 2 |
| chr16_+_19916292 | 1.57 |
ENSMUST00000023509.5
ENSMUST00000232088.2 ENSMUST00000231842.2 |
Klhl24
|
kelch-like 24 |
| chr16_-_93726399 | 1.53 |
ENSMUST00000177648.8
ENSMUST00000142083.2 |
Cldn14
|
claudin 14 |
| chr11_+_118913788 | 1.51 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr12_+_110245662 | 1.49 |
ENSMUST00000097228.5
|
Dio3
|
deiodinase, iodothyronine type III |
| chr11_-_77784922 | 1.49 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr13_+_51254852 | 1.48 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr9_+_78137927 | 1.48 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chr12_+_73333553 | 1.48 |
ENSMUST00000140523.8
ENSMUST00000126488.8 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr1_-_36748985 | 1.48 |
ENSMUST00000043951.10
|
Actr1b
|
ARP1 actin-related protein 1B, centractin beta |
| chr19_+_34268071 | 1.46 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr16_-_4607848 | 1.43 |
ENSMUST00000004173.12
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr3_+_138148846 | 1.43 |
ENSMUST00000005964.7
|
Adh5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr6_+_124785621 | 1.38 |
ENSMUST00000047760.10
|
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr7_-_45264908 | 1.38 |
ENSMUST00000033099.6
|
Fgf21
|
fibroblast growth factor 21 |
| chr10_-_31321938 | 1.37 |
ENSMUST00000000305.7
|
Tpd52l1
|
tumor protein D52-like 1 |
| chr16_-_87229367 | 1.35 |
ENSMUST00000232095.2
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr6_+_124785834 | 1.30 |
ENSMUST00000143040.8
ENSMUST00000052727.5 ENSMUST00000130160.2 |
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr6_+_90442269 | 1.30 |
ENSMUST00000113530.4
|
Klf15
|
Kruppel-like factor 15 |
| chr7_-_130924021 | 1.25 |
ENSMUST00000046611.9
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chr7_+_101859542 | 1.21 |
ENSMUST00000140631.2
ENSMUST00000120879.8 ENSMUST00000146996.8 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chr18_-_80194682 | 1.18 |
ENSMUST00000066743.11
|
Adnp2
|
ADNP homeobox 2 |
| chr2_-_90735171 | 1.18 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr11_-_6394385 | 1.13 |
ENSMUST00000109737.9
|
H2az2
|
H2A.Z histone variant 2 |
| chr7_+_44145987 | 1.09 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr7_+_44146012 | 1.08 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr7_-_7301760 | 1.05 |
ENSMUST00000210061.2
|
Clcn4
|
chloride channel, voltage-sensitive 4 |
| chr19_+_31846154 | 1.05 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr19_-_7183596 | 1.05 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr6_+_90443293 | 1.05 |
ENSMUST00000203607.2
|
Klf15
|
Kruppel-like factor 15 |
| chr9_+_78164402 | 1.03 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chr6_+_17463748 | 0.99 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr5_-_124170305 | 0.98 |
ENSMUST00000040967.9
|
Vps37b
|
vacuolar protein sorting 37B |
| chr4_-_41774097 | 0.98 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr10_+_121575819 | 0.97 |
ENSMUST00000065600.8
ENSMUST00000136432.2 |
BC048403
|
cDNA sequence BC048403 |
| chr11_-_6394352 | 0.96 |
ENSMUST00000093346.6
|
H2az2
|
H2A.Z histone variant 2 |
| chr19_-_7183626 | 0.96 |
ENSMUST00000025679.11
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr17_-_25973288 | 0.96 |
ENSMUST00000075884.8
ENSMUST00000238120.2 ENSMUST00000236137.2 ENSMUST00000237359.2 |
Msln
|
mesothelin |
| chr15_+_68800261 | 0.95 |
ENSMUST00000022954.7
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr9_+_94551929 | 0.92 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr15_+_34238174 | 0.89 |
ENSMUST00000022867.5
ENSMUST00000226627.2 |
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr16_-_94171533 | 0.88 |
ENSMUST00000113910.8
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr4_+_43441939 | 0.88 |
ENSMUST00000060864.13
|
Tesk1
|
testis specific protein kinase 1 |
| chr7_+_44146029 | 0.88 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr1_-_164281344 | 0.87 |
ENSMUST00000193367.2
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr7_-_114235506 | 0.84 |
ENSMUST00000205714.2
ENSMUST00000206853.2 ENSMUST00000205933.2 ENSMUST00000206156.2 ENSMUST00000032907.9 ENSMUST00000032906.11 |
Calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr17_-_14914484 | 0.82 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2 |
| chr1_+_33947250 | 0.81 |
ENSMUST00000183034.5
|
Dst
|
dystonin |
| chr1_-_58625350 | 0.81 |
ENSMUST00000038372.14
ENSMUST00000097724.10 |
Fam126b
|
family with sequence similarity 126, member B |
| chr7_+_43361930 | 0.81 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr6_+_68247469 | 0.80 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr2_+_103858066 | 0.80 |
ENSMUST00000028603.10
|
Fbxo3
|
F-box protein 3 |
| chr1_-_92401459 | 0.79 |
ENSMUST00000185251.2
ENSMUST00000027478.7 |
Ndufa10
|
NADH:ubiquinone oxidoreductase subunit A10 |
| chr7_-_138447933 | 0.77 |
ENSMUST00000118810.2
ENSMUST00000075667.5 ENSMUST00000119664.2 |
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr19_+_43741431 | 0.77 |
ENSMUST00000026199.14
|
Cutc
|
cutC copper transporter |
| chr7_+_138968988 | 0.77 |
ENSMUST00000106098.8
ENSMUST00000026550.14 |
Inpp5a
|
inositol polyphosphate-5-phosphatase A |
| chr10_+_39488930 | 0.76 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr16_-_94171340 | 0.75 |
ENSMUST00000138514.2
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr8_+_95113066 | 0.75 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr4_-_151129020 | 0.74 |
ENSMUST00000103204.11
|
Per3
|
period circadian clock 3 |
| chr19_+_43741513 | 0.73 |
ENSMUST00000112047.10
|
Cutc
|
cutC copper transporter |
| chr15_+_25910510 | 0.73 |
ENSMUST00000228600.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr6_+_67873135 | 0.72 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr16_-_94171556 | 0.71 |
ENSMUST00000113906.9
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr7_-_44145830 | 0.71 |
ENSMUST00000118515.9
ENSMUST00000138328.3 ENSMUST00000239015.2 ENSMUST00000118808.9 |
Emc10
|
ER membrane protein complex subunit 10 |
| chr10_-_43934774 | 0.69 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr11_+_69214895 | 0.69 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chr2_+_3514071 | 0.68 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr17_+_44112679 | 0.67 |
ENSMUST00000229744.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr10_-_128657445 | 0.67 |
ENSMUST00000217685.2
ENSMUST00000026409.5 ENSMUST00000219215.2 ENSMUST00000219524.2 |
Ormdl2
|
ORM1-like 2 (S. cerevisiae) |
| chr7_-_133384449 | 0.65 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr19_-_57170725 | 0.64 |
ENSMUST00000133369.2
|
Ablim1
|
actin-binding LIM protein 1 |
| chr4_-_115911053 | 0.63 |
ENSMUST00000030475.3
|
Nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr17_+_34863738 | 0.63 |
ENSMUST00000036720.9
|
Fkbpl
|
FK506 binding protein-like |
| chr8_-_91860576 | 0.63 |
ENSMUST00000120213.9
ENSMUST00000109609.9 |
Aktip
|
AKT interacting protein |
| chr2_+_132689640 | 0.62 |
ENSMUST00000124836.8
ENSMUST00000154160.2 |
Crls1
|
cardiolipin synthase 1 |
| chr3_-_75177378 | 0.62 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr11_+_110888313 | 0.62 |
ENSMUST00000106635.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr11_-_58425662 | 0.60 |
ENSMUST00000213188.3
|
Olfr330
|
olfactory receptor 330 |
| chr16_-_33916354 | 0.60 |
ENSMUST00000114973.9
ENSMUST00000232157.2 ENSMUST00000114964.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr4_-_119395966 | 0.59 |
ENSMUST00000079611.13
|
Frg2f1
|
FSHD region gene 2 family member 1 |
| chr3_-_142101339 | 0.59 |
ENSMUST00000198381.5
ENSMUST00000090134.12 ENSMUST00000196908.5 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr4_+_40269563 | 0.58 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr19_+_28941292 | 0.58 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr11_+_98828495 | 0.57 |
ENSMUST00000107475.9
ENSMUST00000068133.10 |
Rara
|
retinoic acid receptor, alpha |
| chr10_+_88566918 | 0.57 |
ENSMUST00000116234.9
|
Arl1
|
ADP-ribosylation factor-like 1 |
| chr16_+_43330630 | 0.57 |
ENSMUST00000114695.3
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_+_28910393 | 0.56 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr11_+_69214883 | 0.56 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr2_-_104324035 | 0.55 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr12_-_115611981 | 0.55 |
ENSMUST00000103540.3
ENSMUST00000199266.2 |
Ighv8-12
|
immunoglobulin heavy variable V8-12 |
| chrX_-_153999333 | 0.55 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr9_+_123921573 | 0.55 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr7_+_130294262 | 0.54 |
ENSMUST00000033141.7
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr11_-_83959175 | 0.54 |
ENSMUST00000100705.11
|
Dusp14
|
dual specificity phosphatase 14 |
| chr6_-_97436223 | 0.54 |
ENSMUST00000113359.8
|
Frmd4b
|
FERM domain containing 4B |
| chr8_-_91860655 | 0.53 |
ENSMUST00000125257.3
|
Aktip
|
AKT interacting protein |
| chr17_-_15784582 | 0.52 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
| chr1_+_171173252 | 0.50 |
ENSMUST00000006579.5
|
Pfdn2
|
prefoldin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hlf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 21.0 | GO:0097037 | heme export(GO:0097037) |
| 2.8 | 16.8 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.1 | 6.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.8 | 5.5 | GO:2000487 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 1.8 | 5.5 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 1.4 | 5.5 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.2 | 6.2 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.2 | 39.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 1.0 | 3.1 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 1.0 | 4.1 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.9 | 2.8 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.9 | 3.7 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.9 | 2.7 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.7 | 6.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.7 | 2.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.7 | 2.7 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.6 | 1.7 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.6 | 30.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.6 | 9.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 3.6 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 1.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.5 | 4.4 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.5 | 1.4 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.5 | 4.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.5 | 13.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 4.7 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.4 | 12.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 2.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.4 | 1.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.4 | 5.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 4.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 2.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.3 | 0.9 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.3 | 0.8 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.3 | 1.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.2 | 3.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 2.7 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.2 | 2.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 9.5 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.2 | 1.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 1.6 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 1.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 0.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 1.7 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.2 | 0.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 2.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.2 | 2.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 0.7 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 | 1.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.2 | 1.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 1.5 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.1 | 0.4 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 11.0 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 4.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.8 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.5 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.6 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.6 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.3 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.7 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 6.3 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.5 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.3 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 | 0.9 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 6.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.6 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 2.3 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 2.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 2.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 4.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 2.0 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.8 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 2.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.0 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.6 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.7 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.4 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 1.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.0 | 0.8 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.7 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.0 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.0 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 1.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.0 | GO:0015881 | creatine transport(GO:0015881) |
| 0.0 | 0.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 1.3 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 21.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.6 | 4.7 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.1 | 16.8 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.0 | 4.8 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.6 | 15.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 1.6 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.3 | 3.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 2.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 2.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 15.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.7 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.1 | 2.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.8 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.8 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 4.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 20.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 24.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 9.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 8.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 3.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 35.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 9.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 3.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.8 | GO:0043296 | apical junction complex(GO:0043296) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 14.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.6 | 4.7 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 1.4 | 30.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.4 | 9.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.4 | 4.1 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 1.4 | 5.5 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 1.4 | 5.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 1.1 | 5.5 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 1.0 | 6.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.0 | 37.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.9 | 2.8 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.9 | 2.7 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.8 | 3.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.7 | 2.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.6 | 16.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.6 | 4.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.5 | 2.7 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.5 | 2.7 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.5 | 3.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 3.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 4.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.4 | 3.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 2.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 11.0 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.4 | 1.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.3 | 5.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 4.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 2.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 1.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.3 | 10.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 1.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 2.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 1.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 9.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 7.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 23.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 2.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.2 | 4.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 6.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 0.6 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.2 | 2.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 2.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 2.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 7.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.4 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 1.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 2.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 4.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 2.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 3.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 3.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 4.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.6 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 4.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 3.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 15.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 5.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 5.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 13.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 14.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.7 | 9.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 17.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 4.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 7.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 39.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 5.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 0.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 3.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 4.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 6.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 5.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 4.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 5.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |