Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
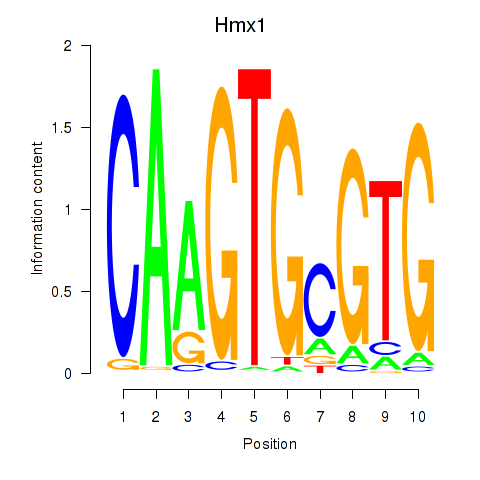
Results for Hmx1
Z-value: 1.95
Transcription factors associated with Hmx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx1
|
ENSMUSG00000067438.6 | H6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx1 | mm39_v1_chr5_+_35546363_35546461 | -0.19 | 2.7e-01 | Click! |
Activity profile of Hmx1 motif
Sorted Z-values of Hmx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_79539063 | 23.90 |
ENSMUST00000022595.8
|
Rgcc
|
regulator of cell cycle |
| chr14_-_56339915 | 23.77 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr7_+_18618605 | 22.18 |
ENSMUST00000032573.8
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr2_+_162916551 | 21.94 |
ENSMUST00000142729.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr4_+_120523758 | 21.28 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr13_+_91609169 | 20.75 |
ENSMUST00000004094.15
ENSMUST00000042122.15 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr11_+_11636213 | 20.56 |
ENSMUST00000076700.11
ENSMUST00000048122.13 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr12_+_109425769 | 17.89 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr6_+_30738043 | 16.89 |
ENSMUST00000163949.9
ENSMUST00000124665.3 |
Mest
|
mesoderm specific transcript |
| chr13_+_91609264 | 14.58 |
ENSMUST00000231481.2
|
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr13_+_45660905 | 13.72 |
ENSMUST00000000260.13
|
Gmpr
|
guanosine monophosphate reductase |
| chrX_-_7537580 | 13.62 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr17_-_34109513 | 13.59 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr11_-_46203047 | 12.93 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr9_+_108437485 | 12.80 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr8_+_106024294 | 11.19 |
ENSMUST00000015003.10
|
E2f4
|
E2F transcription factor 4 |
| chr1_-_75482975 | 10.47 |
ENSMUST00000113567.10
ENSMUST00000113565.3 |
Obsl1
|
obscurin-like 1 |
| chr11_+_61847589 | 10.18 |
ENSMUST00000201671.4
ENSMUST00000202178.4 |
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr6_+_34453142 | 9.74 |
ENSMUST00000045372.6
ENSMUST00000138668.2 ENSMUST00000139067.2 |
Bpgm
|
2,3-bisphosphoglycerate mutase |
| chr4_-_152561896 | 9.57 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chrX_-_133709733 | 9.55 |
ENSMUST00000035559.11
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr1_+_135656885 | 9.22 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr15_-_78456898 | 8.99 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chrX_-_73290140 | 8.96 |
ENSMUST00000101454.9
ENSMUST00000033699.13 |
Flna
|
filamin, alpha |
| chr6_+_90596123 | 8.72 |
ENSMUST00000032177.10
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr2_+_129040677 | 8.59 |
ENSMUST00000028880.10
|
Slc20a1
|
solute carrier family 20, member 1 |
| chrX_+_9138995 | 8.48 |
ENSMUST00000015486.7
|
Xk
|
X-linked Kx blood group |
| chr7_-_125968653 | 8.32 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
| chr5_+_121358254 | 8.06 |
ENSMUST00000042614.13
|
Hectd4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr11_+_69856222 | 8.04 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chrX_-_73289970 | 7.92 |
ENSMUST00000130007.8
|
Flna
|
filamin, alpha |
| chr1_-_181669891 | 7.61 |
ENSMUST00000193028.2
ENSMUST00000191878.6 ENSMUST00000005003.12 |
Lbr
|
lamin B receptor |
| chr11_+_61847622 | 7.59 |
ENSMUST00000202389.4
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr4_+_134195631 | 7.16 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr17_+_27136065 | 7.04 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr7_-_48530777 | 6.94 |
ENSMUST00000058745.15
|
E2f8
|
E2F transcription factor 8 |
| chr6_-_91093766 | 6.61 |
ENSMUST00000113509.2
ENSMUST00000032179.14 |
Nup210
|
nucleoporin 210 |
| chr18_-_62313019 | 6.41 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr2_+_130116344 | 6.16 |
ENSMUST00000103198.11
|
Nop56
|
NOP56 ribonucleoprotein |
| chr17_+_34811217 | 6.11 |
ENSMUST00000038149.13
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr3_-_89325594 | 6.09 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr2_+_152950388 | 6.02 |
ENSMUST00000189688.2
ENSMUST00000109799.8 ENSMUST00000003370.14 |
Hck
|
hemopoietic cell kinase |
| chr2_+_130116357 | 5.69 |
ENSMUST00000136621.9
ENSMUST00000141872.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr15_-_79718423 | 5.57 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr11_-_102210568 | 5.39 |
ENSMUST00000173870.8
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr11_-_109364424 | 5.30 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr13_-_32522548 | 5.15 |
ENSMUST00000041859.9
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr6_-_127128007 | 5.10 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr2_+_32766126 | 5.09 |
ENSMUST00000028135.15
|
Niban2
|
niban apoptosis regulator 2 |
| chr9_-_107474221 | 4.95 |
ENSMUST00000238519.2
|
Lsmem2
|
leucine-rich single-pass membrane protein 2 |
| chr8_+_121262528 | 4.47 |
ENSMUST00000120493.8
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chrX_+_49930311 | 4.44 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr11_-_102209767 | 4.33 |
ENSMUST00000174302.8
ENSMUST00000178839.8 ENSMUST00000006754.14 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr10_-_76797622 | 4.26 |
ENSMUST00000001148.11
ENSMUST00000105411.9 |
Pcbp3
|
poly(rC) binding protein 3 |
| chr3_-_107992662 | 4.03 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr2_-_92201342 | 3.91 |
ENSMUST00000176810.8
ENSMUST00000090582.11 ENSMUST00000068586.13 |
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr11_+_44409775 | 3.89 |
ENSMUST00000019333.10
|
Rnf145
|
ring finger protein 145 |
| chr11_+_68582731 | 3.87 |
ENSMUST00000102611.10
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr6_-_127127959 | 3.85 |
ENSMUST00000201637.2
|
Ccnd2
|
cyclin D2 |
| chr15_-_79571977 | 3.84 |
ENSMUST00000023061.7
|
Josd1
|
Josephin domain containing 1 |
| chr14_-_56499690 | 3.82 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr2_-_156833932 | 3.80 |
ENSMUST00000109558.2
ENSMUST00000069600.13 ENSMUST00000072298.13 |
Ndrg3
|
N-myc downstream regulated gene 3 |
| chr13_-_38842967 | 3.78 |
ENSMUST00000001757.9
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr8_+_121264161 | 3.70 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr13_-_55477535 | 3.69 |
ENSMUST00000021941.8
|
Mxd3
|
Max dimerization protein 3 |
| chr14_-_54924382 | 3.67 |
ENSMUST00000022793.15
ENSMUST00000111484.9 |
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr11_+_68582923 | 3.66 |
ENSMUST00000018887.15
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr15_-_98507913 | 3.59 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr15_+_59246134 | 3.58 |
ENSMUST00000227173.2
ENSMUST00000079703.11 |
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr9_-_42035560 | 3.53 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr7_-_83384711 | 3.51 |
ENSMUST00000001792.12
|
Il16
|
interleukin 16 |
| chr13_+_55517545 | 3.44 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr7_-_125760164 | 3.42 |
ENSMUST00000164741.2
|
Xpo6
|
exportin 6 |
| chr4_+_129407374 | 3.42 |
ENSMUST00000062356.7
|
Marcksl1
|
MARCKS-like 1 |
| chr11_-_116001037 | 3.35 |
ENSMUST00000106441.8
ENSMUST00000021120.6 |
Trim47
|
tripartite motif-containing 47 |
| chr11_-_61384998 | 3.30 |
ENSMUST00000101085.9
ENSMUST00000079080.13 ENSMUST00000108714.2 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr17_+_35413415 | 3.29 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr11_+_23234644 | 3.27 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr6_-_124710030 | 3.22 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr18_+_75953244 | 3.17 |
ENSMUST00000058997.15
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr5_-_134643805 | 3.11 |
ENSMUST00000202085.2
ENSMUST00000036362.13 ENSMUST00000077636.8 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr17_-_34043320 | 2.96 |
ENSMUST00000173879.8
ENSMUST00000166693.3 ENSMUST00000173019.8 |
Rps28
|
ribosomal protein S28 |
| chr12_+_112978051 | 2.94 |
ENSMUST00000223502.2
ENSMUST00000084891.5 ENSMUST00000220541.2 |
Pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr6_-_127127993 | 2.92 |
ENSMUST00000201066.2
|
Ccnd2
|
cyclin D2 |
| chr9_-_110483210 | 2.91 |
ENSMUST00000196488.5
ENSMUST00000133191.8 ENSMUST00000167320.8 |
Nbeal2
|
neurobeachin-like 2 |
| chr16_+_18695787 | 2.90 |
ENSMUST00000120532.9
ENSMUST00000004222.14 |
Hira
|
histone cell cycle regulator |
| chr17_-_48144094 | 2.89 |
ENSMUST00000131971.2
ENSMUST00000129360.2 ENSMUST00000113280.8 ENSMUST00000132125.8 |
Mdfi
|
MyoD family inhibitor |
| chr16_+_49519561 | 2.83 |
ENSMUST00000046777.11
ENSMUST00000142682.9 |
Ift57
|
intraflagellar transport 57 |
| chr17_-_48144181 | 2.82 |
ENSMUST00000152455.8
ENSMUST00000035375.14 |
Mdfi
|
MyoD family inhibitor |
| chr16_+_10652910 | 2.75 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr9_-_21223551 | 2.69 |
ENSMUST00000003397.9
ENSMUST00000213250.2 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr17_-_12988492 | 2.64 |
ENSMUST00000024599.14
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr5_-_38719025 | 2.59 |
ENSMUST00000005234.13
|
Wdr1
|
WD repeat domain 1 |
| chr2_+_118756973 | 2.50 |
ENSMUST00000099546.6
ENSMUST00000110837.2 |
Chst14
|
carbohydrate sulfotransferase 14 |
| chr6_-_124710084 | 2.47 |
ENSMUST00000112484.10
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_-_115024807 | 2.47 |
ENSMUST00000106561.8
ENSMUST00000051264.14 ENSMUST00000106562.3 |
Cd300lf
|
CD300 molecule like family member F |
| chr5_+_100666278 | 2.46 |
ENSMUST00000151414.8
|
Cops4
|
COP9 signalosome subunit 4 |
| chr5_+_100666175 | 2.44 |
ENSMUST00000045993.15
|
Cops4
|
COP9 signalosome subunit 4 |
| chr15_+_79025523 | 2.42 |
ENSMUST00000040077.8
|
Polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr19_-_4665668 | 2.42 |
ENSMUST00000113822.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr19_-_4665509 | 2.41 |
ENSMUST00000053597.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr6_+_29433247 | 2.40 |
ENSMUST00000101617.9
ENSMUST00000065090.8 |
Flnc
|
filamin C, gamma |
| chr2_-_25162347 | 2.37 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr6_-_137146708 | 2.22 |
ENSMUST00000117919.8
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chrX_+_20291927 | 2.18 |
ENSMUST00000115384.9
|
Jade3
|
jade family PHD finger 3 |
| chr14_-_54924062 | 2.14 |
ENSMUST00000147714.8
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr9_+_32607301 | 2.12 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chrX_-_73416824 | 2.11 |
ENSMUST00000178691.2
ENSMUST00000114146.8 |
Ubl4a
Slc10a3
|
ubiquitin-like 4A solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr9_+_106048116 | 2.10 |
ENSMUST00000020490.13
|
Wdr82
|
WD repeat domain containing 82 |
| chrX_-_73416869 | 2.08 |
ENSMUST00000073067.11
ENSMUST00000037967.6 |
Slc10a3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr4_-_94444975 | 2.06 |
ENSMUST00000030313.9
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr10_+_77095052 | 2.05 |
ENSMUST00000020493.9
|
Pofut2
|
protein O-fucosyltransferase 2 |
| chr7_+_118311740 | 2.04 |
ENSMUST00000106557.8
|
Ccp110
|
centriolar coiled coil protein 110 |
| chr2_-_73284262 | 2.02 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr11_-_116734275 | 1.96 |
ENSMUST00000047616.10
|
Jmjd6
|
jumonji domain containing 6 |
| chr11_+_77873535 | 1.95 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr1_-_156546600 | 1.93 |
ENSMUST00000122424.8
ENSMUST00000086153.8 |
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr18_-_24736848 | 1.93 |
ENSMUST00000070726.10
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr6_-_87312743 | 1.92 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr15_+_8138805 | 1.91 |
ENSMUST00000230017.2
|
Nup155
|
nucleoporin 155 |
| chr10_-_61109188 | 1.88 |
ENSMUST00000092486.11
|
Adamts14
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
| chr3_+_102965601 | 1.87 |
ENSMUST00000029445.13
ENSMUST00000200069.5 |
Nras
|
neuroblastoma ras oncogene |
| chr11_-_69749549 | 1.83 |
ENSMUST00000001626.10
ENSMUST00000108626.8 |
Tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr6_-_137146677 | 1.81 |
ENSMUST00000119610.4
ENSMUST00000149100.3 |
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr12_+_70984631 | 1.75 |
ENSMUST00000021479.6
|
Actr10
|
ARP10 actin-related protein 10 |
| chr14_-_56812839 | 1.69 |
ENSMUST00000225951.2
|
Cenpj
|
centromere protein J |
| chr9_-_21223631 | 1.69 |
ENSMUST00000115433.11
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr11_-_5015275 | 1.66 |
ENSMUST00000109895.2
ENSMUST00000152257.2 ENSMUST00000037146.10 ENSMUST00000056649.13 |
Gas2l1
|
growth arrest-specific 2 like 1 |
| chr14_-_56472102 | 1.52 |
ENSMUST00000015585.4
|
Gzmc
|
granzyme C |
| chr15_-_74581384 | 1.52 |
ENSMUST00000050234.4
|
Jrk
|
jerky |
| chrX_-_99670174 | 1.48 |
ENSMUST00000015812.12
|
Pdzd11
|
PDZ domain containing 11 |
| chrX_+_158038778 | 1.48 |
ENSMUST00000126686.8
ENSMUST00000033671.13 |
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr3_-_92393193 | 1.48 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr1_-_152642073 | 1.46 |
ENSMUST00000111857.3
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr15_+_59246080 | 1.45 |
ENSMUST00000168722.3
|
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr15_+_78312851 | 1.44 |
ENSMUST00000159771.8
|
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr1_-_127605660 | 1.41 |
ENSMUST00000160616.8
|
Tmem163
|
transmembrane protein 163 |
| chr3_-_88455556 | 1.37 |
ENSMUST00000131775.2
ENSMUST00000008745.13 |
Rab25
|
RAB25, member RAS oncogene family |
| chr3_-_107993906 | 1.36 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr7_-_117842787 | 1.33 |
ENSMUST00000032891.15
|
Smg1
|
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) |
| chr2_+_25162487 | 1.31 |
ENSMUST00000028341.11
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr11_+_88864753 | 1.31 |
ENSMUST00000036649.8
|
Coil
|
coilin |
| chr6_-_40413056 | 1.27 |
ENSMUST00000039008.10
ENSMUST00000101492.10 |
Dennd11
|
DENN domain containing 11 |
| chr1_-_152642032 | 1.26 |
ENSMUST00000111859.8
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr8_+_108162985 | 1.24 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr15_+_78312764 | 1.22 |
ENSMUST00000162517.8
ENSMUST00000166142.10 ENSMUST00000089414.11 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr1_-_143578542 | 1.21 |
ENSMUST00000018337.9
|
Cdc73
|
cell division cycle 73, Paf1/RNA polymerase II complex component |
| chr5_+_124583524 | 1.19 |
ENSMUST00000100709.7
|
Kmt5a
|
lysine methyltransferase 5A |
| chr12_-_46767619 | 1.18 |
ENSMUST00000219886.2
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr7_+_100021425 | 1.16 |
ENSMUST00000098259.11
ENSMUST00000051777.15 |
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr2_+_174169351 | 1.14 |
ENSMUST00000124935.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr15_+_8138757 | 1.11 |
ENSMUST00000163765.3
|
Nup155
|
nucleoporin 155 |
| chr1_-_160134873 | 1.10 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr1_+_180468895 | 1.05 |
ENSMUST00000192725.6
|
Lin9
|
lin-9 homolog (C. elegans) |
| chrX_-_73436293 | 1.04 |
ENSMUST00000114138.8
|
Fam3a
|
family with sequence similarity 3, member A |
| chrX_-_73436609 | 1.04 |
ENSMUST00000015427.13
|
Fam3a
|
family with sequence similarity 3, member A |
| chr19_-_5345423 | 1.04 |
ENSMUST00000235182.2
ENSMUST00000025774.11 |
Sf3b2
|
splicing factor 3b, subunit 2 |
| chr1_+_39574829 | 1.03 |
ENSMUST00000003219.14
|
Cnot11
|
CCR4-NOT transcription complex, subunit 11 |
| chr4_+_109533753 | 0.98 |
ENSMUST00000102724.5
|
Faf1
|
Fas-associated factor 1 |
| chr2_-_181234526 | 0.98 |
ENSMUST00000108789.9
ENSMUST00000153998.2 |
Zfp512b
|
zinc finger protein 512B |
| chr9_-_44646487 | 0.96 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr11_+_62711057 | 0.95 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr2_-_34716083 | 0.93 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr1_+_61017057 | 0.92 |
ENSMUST00000027162.12
ENSMUST00000102827.4 |
Icos
|
inducible T cell co-stimulator |
| chr11_-_94133527 | 0.90 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr19_-_14575395 | 0.89 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr15_+_78294154 | 0.87 |
ENSMUST00000229739.2
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr16_-_97564910 | 0.87 |
ENSMUST00000019386.10
|
Ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr1_+_52047368 | 0.86 |
ENSMUST00000027277.7
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr2_-_92201311 | 0.84 |
ENSMUST00000176774.8
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr17_-_36291087 | 0.83 |
ENSMUST00000055454.14
|
Prr3
|
proline-rich polypeptide 3 |
| chr11_-_32217547 | 0.82 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr5_-_143513732 | 0.81 |
ENSMUST00000100489.4
ENSMUST00000080537.14 |
Rac1
|
Rac family small GTPase 1 |
| chr15_-_79025387 | 0.78 |
ENSMUST00000187550.7
ENSMUST00000188562.7 ENSMUST00000190509.7 ENSMUST00000190730.7 ENSMUST00000190959.7 ENSMUST00000169604.8 ENSMUST00000186053.7 |
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chrX_-_73436709 | 0.77 |
ENSMUST00000114142.8
ENSMUST00000114139.8 |
Fam3a
|
family with sequence similarity 3, member A |
| chr4_+_59626185 | 0.76 |
ENSMUST00000070150.11
ENSMUST00000052420.7 |
E130308A19Rik
|
RIKEN cDNA E130308A19 gene |
| chr2_+_28083105 | 0.75 |
ENSMUST00000100244.10
|
Olfm1
|
olfactomedin 1 |
| chr2_-_130471891 | 0.75 |
ENSMUST00000110262.3
ENSMUST00000028761.5 |
Fastkd5
Ubox5
|
FAST kinase domains 5 U box domain containing 5 |
| chr19_+_8713156 | 0.72 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr15_+_55171138 | 0.70 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr19_-_32365862 | 0.69 |
ENSMUST00000099514.10
|
Sgms1
|
sphingomyelin synthase 1 |
| chr7_+_48438751 | 0.69 |
ENSMUST00000118927.8
ENSMUST00000125280.8 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chr19_-_41969558 | 0.69 |
ENSMUST00000026168.9
ENSMUST00000171561.8 |
Mms19
|
MMS19 cytosolic iron-sulfur assembly component |
| chr1_+_43769750 | 0.68 |
ENSMUST00000027217.9
|
Ecrg4
|
ECRG4 augurin precursor |
| chr10_+_79500387 | 0.68 |
ENSMUST00000020554.8
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr4_+_115595610 | 0.67 |
ENSMUST00000106524.4
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr15_+_86098660 | 0.64 |
ENSMUST00000063414.9
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr2_+_174169492 | 0.63 |
ENSMUST00000156623.8
ENSMUST00000149016.9 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr4_+_62537750 | 0.62 |
ENSMUST00000084521.11
ENSMUST00000107424.2 |
Rgs3
|
regulator of G-protein signaling 3 |
| chr16_-_97412169 | 0.61 |
ENSMUST00000232141.2
ENSMUST00000000395.8 |
Tmprss2
|
transmembrane protease, serine 2 |
| chr17_-_37178079 | 0.60 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr4_+_129878890 | 0.60 |
ENSMUST00000106017.8
ENSMUST00000121049.8 |
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr11_-_48836975 | 0.59 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr2_-_39116284 | 0.57 |
ENSMUST00000204701.3
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr2_+_174169539 | 0.56 |
ENSMUST00000133356.8
ENSMUST00000087871.11 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr6_-_67512768 | 0.53 |
ENSMUST00000058178.6
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr9_+_95836797 | 0.52 |
ENSMUST00000034981.14
ENSMUST00000185633.7 |
Xrn1
|
5'-3' exoribonuclease 1 |
| chr1_+_152642291 | 0.52 |
ENSMUST00000077755.11
ENSMUST00000097536.6 |
Arpc5
|
actin related protein 2/3 complex, subunit 5 |
| chr6_+_108637816 | 0.51 |
ENSMUST00000163617.2
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr15_-_59245998 | 0.50 |
ENSMUST00000022976.6
|
Washc5
|
WASH complex subunit 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 23.9 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 7.4 | 22.2 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 6.9 | 20.6 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 5.6 | 16.9 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 3.4 | 13.6 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 2.4 | 23.8 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 2.3 | 6.9 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 1.8 | 12.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 1.8 | 7.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.6 | 6.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 1.4 | 8.5 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 1.3 | 12.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.2 | 3.6 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 1.1 | 3.3 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 1.1 | 5.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 1.0 | 7.2 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.0 | 9.7 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.9 | 5.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.9 | 3.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) positive regulation of endocytic recycling(GO:2001137) |
| 0.8 | 5.0 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.8 | 2.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.8 | 7.9 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.8 | 3.0 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.7 | 11.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.7 | 5.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.7 | 5.7 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.7 | 10.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.6 | 2.5 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) interleukin-4-mediated signaling pathway(GO:0035771) response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.6 | 2.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.6 | 5.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.6 | 2.9 | GO:1905035 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.6 | 10.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.5 | 5.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.5 | 9.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.5 | 2.9 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.5 | 4.8 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 3.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.4 | 16.1 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.4 | 8.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.4 | 2.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 13.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.4 | 2.9 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.4 | 2.0 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.4 | 8.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.4 | 2.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.3 | 2.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.3 | 3.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.3 | 2.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 3.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 21.9 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.3 | 1.7 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.3 | 11.9 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.3 | 3.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 8.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.3 | 5.7 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.3 | 2.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.3 | 4.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 16.9 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.2 | 1.5 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 3.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 2.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.2 | 6.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 2.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 2.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 0.8 | GO:0002551 | mast cell chemotaxis(GO:0002551) |
| 0.2 | 2.9 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.0 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 1.9 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 1.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 4.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 2.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.9 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.1 | 2.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 16.5 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.1 | 0.6 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 9.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 6.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 3.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.4 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 0.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 1.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 1.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 2.7 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.5 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 0.4 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 1.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.7 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 2.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 3.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 3.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.0 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.6 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 3.1 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.1 | 1.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 6.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 2.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.9 | GO:0031295 | T cell tolerance induction(GO:0002517) T cell costimulation(GO:0031295) |
| 0.0 | 1.2 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.0 | 0.3 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 2.8 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.0 | 0.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0043545 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.4 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 5.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 7.3 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.7 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 3.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 1.0 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.3 | GO:0046039 | GTP metabolic process(GO:0046039) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 38.8 | GO:0031523 | Myb complex(GO:0031523) |
| 4.6 | 13.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 3.2 | 9.6 | GO:1990031 | pinceau fiber(GO:1990031) |
| 1.5 | 11.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.3 | 11.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 1.0 | 7.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.0 | 20.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.9 | 2.6 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.8 | 3.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.6 | 5.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.5 | 13.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.5 | 5.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.5 | 5.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 3.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 10.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.4 | 7.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) nuclear lamina(GO:0005652) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.4 | 6.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 22.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 3.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 8.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 3.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 2.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 2.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 0.7 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 13.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 1.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 3.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 2.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 4.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 1.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 12.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 2.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 5.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 4.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 6.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 3.7 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 5.3 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 4.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 8.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 24.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 3.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 10.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 3.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.2 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 0.3 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 2.3 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 18.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.7 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.0 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 3.7 | 22.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 3.2 | 9.7 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 2.8 | 16.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 2.1 | 12.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 1.7 | 8.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.6 | 19.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.6 | 9.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 1.6 | 6.4 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 1.0 | 6.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.7 | 4.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.7 | 7.6 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.7 | 11.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.6 | 2.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.6 | 23.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.5 | 20.6 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.5 | 5.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 4.8 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.5 | 10.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 4.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 3.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.5 | 2.5 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.5 | 2.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 13.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.4 | 5.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 13.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.4 | 9.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.4 | 3.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 0.7 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.3 | 2.0 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.3 | 4.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.3 | 7.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 0.9 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.3 | 20.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 11.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 4.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 35.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.3 | 3.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 2.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 3.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 2.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.2 | 2.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 1.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 2.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 5.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 8.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 3.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 3.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 3.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 3.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 5.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.8 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 3.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 6.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 15.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 3.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 5.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 5.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.2 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 2.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.3 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 10.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 2.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 3.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 3.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 8.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 7.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 24.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 16.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.3 | 21.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.3 | 15.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 17.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 19.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 21.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 12.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 6.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 3.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 3.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 6.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 1.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 3.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 5.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 34.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 1.1 | 23.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.8 | 19.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.7 | 11.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.6 | 9.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.5 | 17.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 13.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 17.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.4 | 6.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 17.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 3.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 3.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 4.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 8.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 5.7 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.3 | 9.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 6.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 10.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 1.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 6.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 9.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 4.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 3.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 4.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 3.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.5 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 2.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 5.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |