Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxa1
Z-value: 0.56
Transcription factors associated with Hoxa1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa1
|
ENSMUSG00000029844.10 | homeobox A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa1 | mm39_v1_chr6_-_52135261_52135332 | -0.52 | 1.2e-03 | Click! |
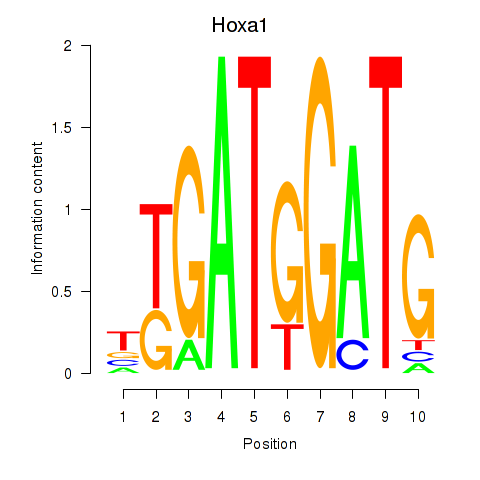
Activity profile of Hoxa1 motif
Sorted Z-values of Hoxa1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_118400418 | 3.41 |
ENSMUST00000173522.8
ENSMUST00000174450.2 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr19_-_7943365 | 3.31 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr13_-_12355604 | 2.31 |
ENSMUST00000168193.8
ENSMUST00000064204.14 |
Actn2
|
actinin alpha 2 |
| chr16_+_38722666 | 1.84 |
ENSMUST00000023478.8
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr16_-_10360893 | 1.72 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr2_-_103315483 | 1.51 |
ENSMUST00000028610.10
|
Cat
|
catalase |
| chr18_-_56695288 | 1.24 |
ENSMUST00000170309.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr18_-_56695333 | 1.18 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr6_-_21851827 | 1.18 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr18_-_56695259 | 1.18 |
ENSMUST00000171844.3
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr6_-_136852792 | 0.93 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr10_-_125225298 | 0.90 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr15_+_9140614 | 0.89 |
ENSMUST00000227556.3
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr11_+_86375441 | 0.81 |
ENSMUST00000020827.7
|
Rnft1
|
ring finger protein, transmembrane 1 |
| chr19_-_47452557 | 0.77 |
ENSMUST00000111800.4
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr16_+_22926162 | 0.76 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr2_+_152804405 | 0.75 |
ENSMUST00000099197.9
ENSMUST00000103155.10 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr6_-_83654789 | 0.75 |
ENSMUST00000037882.8
|
Cd207
|
CD207 antigen |
| chr13_+_41040657 | 0.64 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr14_+_70314652 | 0.64 |
ENSMUST00000035908.3
|
Egr3
|
early growth response 3 |
| chr6_-_93889483 | 0.60 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr17_-_25300112 | 0.59 |
ENSMUST00000024984.7
|
Tmem204
|
transmembrane protein 204 |
| chr4_+_43669266 | 0.55 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr19_-_3625698 | 0.55 |
ENSMUST00000172362.3
ENSMUST00000025846.16 ENSMUST00000226109.2 ENSMUST00000113997.9 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr2_-_104324035 | 0.53 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr15_-_53209513 | 0.52 |
ENSMUST00000077273.9
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr1_+_153750081 | 0.50 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chr5_+_42225303 | 0.49 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr1_-_157240138 | 0.49 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
| chr11_+_4833186 | 0.48 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chr7_-_103320398 | 0.44 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr4_-_145041710 | 0.42 |
ENSMUST00000030339.13
|
Tnfrsf8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr11_-_118460736 | 0.41 |
ENSMUST00000136551.3
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr2_+_180829040 | 0.41 |
ENSMUST00000016488.13
ENSMUST00000108841.2 |
Ppdpf
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr3_-_58433313 | 0.41 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr1_-_43866910 | 0.39 |
ENSMUST00000153317.6
ENSMUST00000128261.2 ENSMUST00000126008.8 ENSMUST00000139451.8 |
Uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr4_-_143026068 | 0.38 |
ENSMUST00000030317.14
|
Pdpn
|
podoplanin |
| chr6_+_95094721 | 0.37 |
ENSMUST00000032107.10
ENSMUST00000119582.3 |
Kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chrX_+_109857866 | 0.36 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr7_+_123061535 | 0.36 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr10_-_33500583 | 0.35 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr5_-_138981842 | 0.34 |
ENSMUST00000110897.8
|
Pdgfa
|
platelet derived growth factor, alpha |
| chr11_-_40646090 | 0.33 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr5_+_88731386 | 0.32 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr4_-_148021159 | 0.31 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr9_+_121727421 | 0.31 |
ENSMUST00000214340.2
ENSMUST00000050327.5 |
Ackr2
|
atypical chemokine receptor 2 |
| chr15_-_95426419 | 0.30 |
ENSMUST00000229933.2
ENSMUST00000166170.9 |
Nell2
|
NEL-like 2 |
| chr9_+_78303059 | 0.30 |
ENSMUST00000113367.2
|
Ddx43
|
DEAD box helicase 43 |
| chr17_+_26036893 | 0.30 |
ENSMUST00000235694.2
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr17_-_90763300 | 0.29 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr15_+_89095724 | 0.28 |
ENSMUST00000227951.2
ENSMUST00000226221.2 ENSMUST00000238818.2 ENSMUST00000228284.2 |
Ppp6r2
|
protein phosphatase 6, regulatory subunit 2 |
| chr3_+_97920819 | 0.28 |
ENSMUST00000079812.8
|
Notch2
|
notch 2 |
| chr5_+_88731366 | 0.28 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr18_+_37108994 | 0.26 |
ENSMUST00000193984.2
|
Gm37388
|
predicted gene, 37388 |
| chr2_+_158217558 | 0.26 |
ENSMUST00000109488.8
|
Snhg11
|
small nucleolar RNA host gene 11 |
| chr2_-_63014622 | 0.25 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr1_+_74700952 | 0.25 |
ENSMUST00000129890.8
|
Ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr3_-_7678785 | 0.25 |
ENSMUST00000194279.6
|
Il7
|
interleukin 7 |
| chr5_+_137059522 | 0.25 |
ENSMUST00000187382.2
|
Vgf
|
VGF nerve growth factor inducible |
| chr1_+_173964312 | 0.24 |
ENSMUST00000053941.4
|
Olfr424
|
olfactory receptor 424 |
| chr4_-_106657446 | 0.24 |
ENSMUST00000148688.2
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr3_-_49711765 | 0.24 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr4_-_143026033 | 0.24 |
ENSMUST00000119654.2
|
Pdpn
|
podoplanin |
| chr15_+_100262423 | 0.24 |
ENSMUST00000175683.8
ENSMUST00000177211.2 |
Higd1c
|
HIG1 domain family, member 1C |
| chr5_+_137059127 | 0.24 |
ENSMUST00000041543.9
ENSMUST00000186451.2 |
Vgf
|
VGF nerve growth factor inducible |
| chr13_+_63387827 | 0.22 |
ENSMUST00000222929.2
|
Aopep
|
aminopeptidase O |
| chr2_-_69715899 | 0.22 |
ENSMUST00000060447.13
|
Mettl5
|
methyltransferase like 5 |
| chr6_-_88423464 | 0.22 |
ENSMUST00000204459.3
ENSMUST00000203213.3 ENSMUST00000205179.3 ENSMUST00000165242.4 |
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr2_+_32127309 | 0.22 |
ENSMUST00000123740.3
|
Pomt1
|
protein-O-mannosyltransferase 1 |
| chr9_+_15150341 | 0.22 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr6_+_15727798 | 0.22 |
ENSMUST00000128849.3
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr14_+_66581745 | 0.21 |
ENSMUST00000152093.8
ENSMUST00000074523.13 |
Stmn4
|
stathmin-like 4 |
| chr14_+_70314727 | 0.21 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chrX_-_7999009 | 0.21 |
ENSMUST00000130832.8
ENSMUST00000033506.13 ENSMUST00000115623.8 ENSMUST00000153839.2 |
Wdr13
|
WD repeat domain 13 |
| chr15_+_74388044 | 0.21 |
ENSMUST00000042035.16
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr19_+_42040681 | 0.21 |
ENSMUST00000164518.4
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
| chr3_-_49711706 | 0.21 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr7_+_73040908 | 0.20 |
ENSMUST00000128471.2
ENSMUST00000139780.3 |
Rgma
|
repulsive guidance molecule family member A |
| chr14_+_66581818 | 0.20 |
ENSMUST00000118426.8
ENSMUST00000121955.8 ENSMUST00000120229.8 ENSMUST00000134440.2 |
Stmn4
|
stathmin-like 4 |
| chr10_+_81464536 | 0.20 |
ENSMUST00000129622.2
|
Ankrd24
|
ankyrin repeat domain 24 |
| chr7_+_123061497 | 0.20 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr8_+_11362805 | 0.19 |
ENSMUST00000033899.14
|
Col4a2
|
collagen, type IV, alpha 2 |
| chr10_+_36383008 | 0.19 |
ENSMUST00000168572.8
|
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr3_-_88669551 | 0.19 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr13_+_63387870 | 0.19 |
ENSMUST00000159152.3
ENSMUST00000221820.2 |
Aopep
|
aminopeptidase O |
| chr2_-_63014514 | 0.18 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr7_+_141048191 | 0.18 |
ENSMUST00000211071.2
|
Cd151
|
CD151 antigen |
| chr9_+_59496571 | 0.18 |
ENSMUST00000121266.8
ENSMUST00000118164.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr4_-_148021217 | 0.17 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr15_+_98953776 | 0.17 |
ENSMUST00000229268.2
|
Prph
|
peripherin |
| chr13_+_31740117 | 0.16 |
ENSMUST00000042118.11
|
Foxq1
|
forkhead box Q1 |
| chr3_+_58913234 | 0.15 |
ENSMUST00000040846.15
|
Med12l
|
mediator complex subunit 12-like |
| chr3_-_57202546 | 0.15 |
ENSMUST00000196506.2
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr3_-_89000591 | 0.15 |
ENSMUST00000090929.12
ENSMUST00000052539.13 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr1_+_153541020 | 0.15 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr16_-_89284830 | 0.15 |
ENSMUST00000072280.5
|
Krtap8-1
|
keratin associated protein 8-1 |
| chr9_-_4796217 | 0.14 |
ENSMUST00000027020.13
ENSMUST00000163309.2 |
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr17_+_87224776 | 0.14 |
ENSMUST00000042172.7
|
Tmem247
|
transmembrane protein 247 |
| chr3_-_7678796 | 0.14 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr7_+_119289249 | 0.13 |
ENSMUST00000047045.10
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr12_-_8351954 | 0.13 |
ENSMUST00000220073.2
ENSMUST00000037313.6 |
Gdf7
|
growth differentiation factor 7 |
| chr17_-_35023521 | 0.13 |
ENSMUST00000025223.9
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr14_+_56813060 | 0.13 |
ENSMUST00000161553.2
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr18_+_56695515 | 0.13 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr10_+_81068980 | 0.12 |
ENSMUST00000144087.2
ENSMUST00000117798.8 |
Zfr2
|
zinc finger RNA binding protein 2 |
| chr1_+_153767478 | 0.12 |
ENSMUST00000050660.6
|
Teddm1a
|
transmembrane epididymal protein 1A |
| chr11_-_5848771 | 0.11 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr9_-_4796142 | 0.11 |
ENSMUST00000063508.15
|
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr8_-_11362731 | 0.11 |
ENSMUST00000033898.10
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr9_+_58036345 | 0.11 |
ENSMUST00000085677.9
|
Stra6
|
stimulated by retinoic acid gene 6 |
| chr15_-_78272957 | 0.11 |
ENSMUST00000169575.2
|
Tex33
|
testis expressed 33 |
| chr7_+_139902515 | 0.10 |
ENSMUST00000078103.3
|
Olfr525
|
olfactory receptor 525 |
| chr2_+_119803180 | 0.09 |
ENSMUST00000066058.8
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr16_+_16714333 | 0.09 |
ENSMUST00000027373.12
ENSMUST00000232247.2 |
Ppm1f
|
protein phosphatase 1F (PP2C domain containing) |
| chr11_+_58556387 | 0.09 |
ENSMUST00000072030.4
|
Olfr322
|
olfactory receptor 322 |
| chr10_+_81464370 | 0.08 |
ENSMUST00000123896.8
ENSMUST00000153573.2 ENSMUST00000119336.8 |
Ankrd24
|
ankyrin repeat domain 24 |
| chr11_-_69439934 | 0.08 |
ENSMUST00000108659.2
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr1_+_11063678 | 0.08 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr18_+_82929451 | 0.08 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr4_+_138503046 | 0.07 |
ENSMUST00000030528.9
|
Pla2g2d
|
phospholipase A2, group IID |
| chr2_-_152793607 | 0.07 |
ENSMUST00000109811.4
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr18_+_82929037 | 0.07 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr9_+_58036759 | 0.06 |
ENSMUST00000133287.8
|
Stra6
|
stimulated by retinoic acid gene 6 |
| chr2_-_152793469 | 0.06 |
ENSMUST00000037715.7
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr9_+_58036604 | 0.06 |
ENSMUST00000034880.10
|
Stra6
|
stimulated by retinoic acid gene 6 |
| chr6_-_52168675 | 0.05 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr5_+_43673093 | 0.05 |
ENSMUST00000144558.3
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr11_-_65050716 | 0.05 |
ENSMUST00000020855.5
ENSMUST00000108696.7 |
1700086D15Rik
|
RIKEN cDNA 1700086D15 gene |
| chr7_-_102507962 | 0.04 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr2_+_119803230 | 0.02 |
ENSMUST00000229024.2
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr17_+_45866618 | 0.02 |
ENSMUST00000024742.9
ENSMUST00000233929.2 |
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon |
| chr6_+_40302106 | 0.02 |
ENSMUST00000031977.12
|
Agk
|
acylglycerol kinase |
| chr10_+_88721854 | 0.02 |
ENSMUST00000020255.8
|
Slc5a8
|
solute carrier family 5 (iodide transporter), member 8 |
| chr7_+_144391786 | 0.02 |
ENSMUST00000155320.8
|
Fgf3
|
fibroblast growth factor 3 |
| chr15_+_6552270 | 0.02 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr6_+_40419797 | 0.02 |
ENSMUST00000038907.9
ENSMUST00000141490.2 |
Wee2
|
WEE1 homolog 2 (S. pombe) |
| chr8_-_91544021 | 0.01 |
ENSMUST00000209208.2
|
Gm19935
|
predicted gene, 19935 |
| chr10_+_67021509 | 0.01 |
ENSMUST00000173689.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chrX_-_133062677 | 0.01 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr13_+_113429690 | 0.01 |
ENSMUST00000136755.10
|
Cspg4b
|
chondroitin sulfate proteoglycan 4B |
| chr5_-_135601887 | 0.01 |
ENSMUST00000004936.10
ENSMUST00000201401.2 |
Ccl24
|
chemokine (C-C motif) ligand 24 |
| chrX_-_36432482 | 0.00 |
ENSMUST00000046557.6
|
Akap14
|
A kinase (PRKA) anchor protein 14 |
| chr2_+_36342599 | 0.00 |
ENSMUST00000072854.2
|
Olfr340
|
olfactory receptor 340 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0086097 | actin filament uncapping(GO:0051695) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 3.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.6 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.9 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.1 | 1.5 | GO:0009650 | UV protection(GO:0009650) |
| 0.1 | 0.5 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.3 | GO:1990705 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.0 | 0.2 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.8 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.4 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.4 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.4 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.6 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.4 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.6 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 2.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.5 | 2.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 3.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 0.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.2 | 3.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 0.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.9 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.5 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 3.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |