Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
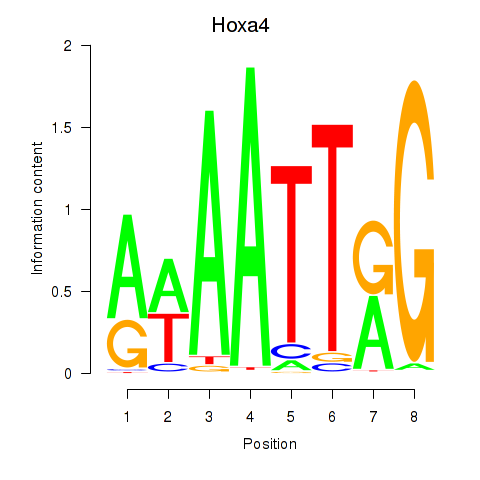
Results for Hoxa4
Z-value: 1.40
Transcription factors associated with Hoxa4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa4
|
ENSMUSG00000000942.11 | homeobox A4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa4 | mm39_v1_chr6_-_52168675_52168733 | -0.64 | 3.0e-05 | Click! |
Activity profile of Hoxa4 motif
Sorted Z-values of Hoxa4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_61259997 | 24.49 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr4_-_60222580 | 22.84 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr4_-_61259801 | 22.45 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr4_-_60697274 | 14.12 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr7_-_19432933 | 11.29 |
ENSMUST00000174355.8
ENSMUST00000172983.8 ENSMUST00000174710.2 ENSMUST00000003066.16 ENSMUST00000174064.9 |
Apoe
|
apolipoprotein E |
| chr5_-_87074380 | 8.52 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr1_-_180021039 | 6.65 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr9_-_50466470 | 6.51 |
ENSMUST00000119103.2
|
Bco2
|
beta-carotene oxygenase 2 |
| chr1_-_180021218 | 5.96 |
ENSMUST00000159914.8
|
Coq8a
|
coenzyme Q8A |
| chr7_+_51528715 | 5.68 |
ENSMUST00000051912.13
|
Gas2
|
growth arrest specific 2 |
| chr10_+_87695117 | 5.59 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr7_+_51528788 | 5.14 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr1_+_74324089 | 4.64 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr10_+_87694924 | 4.51 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr16_+_37400590 | 4.27 |
ENSMUST00000159787.8
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr16_+_22676589 | 4.13 |
ENSMUST00000004574.14
ENSMUST00000178320.2 ENSMUST00000166487.10 |
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr1_+_87998487 | 3.95 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr16_+_37400500 | 3.85 |
ENSMUST00000160847.2
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr11_-_43727071 | 3.74 |
ENSMUST00000167574.2
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr9_-_65792306 | 3.72 |
ENSMUST00000122410.8
ENSMUST00000117083.2 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr6_-_116084810 | 3.58 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr15_+_10249646 | 3.54 |
ENSMUST00000134410.8
|
Prlr
|
prolactin receptor |
| chrM_+_5319 | 3.34 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr12_-_57592907 | 3.23 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr11_-_116080361 | 3.14 |
ENSMUST00000148601.2
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr12_+_59176543 | 3.04 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr11_+_94219046 | 3.02 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr2_-_5680801 | 2.95 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr2_+_70305267 | 2.84 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chr11_+_94218810 | 2.76 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr10_-_126956991 | 2.73 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr5_+_73648368 | 2.56 |
ENSMUST00000113558.8
ENSMUST00000063882.12 |
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr5_+_42225303 | 2.55 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr12_+_59176506 | 2.51 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr19_+_31846154 | 2.43 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr6_+_43242516 | 2.42 |
ENSMUST00000031750.14
|
Arhgef5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_134099840 | 2.42 |
ENSMUST00000030643.3
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chrX_-_105647282 | 2.02 |
ENSMUST00000113480.2
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr5_+_30972067 | 2.00 |
ENSMUST00000200692.4
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr14_+_47069582 | 1.96 |
ENSMUST00000068532.10
|
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr1_-_93406091 | 1.95 |
ENSMUST00000188165.2
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chrM_+_10167 | 1.91 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr1_+_133109059 | 1.85 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr7_+_65343156 | 1.81 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr9_+_7445822 | 1.78 |
ENSMUST00000034497.8
|
Mmp3
|
matrix metallopeptidase 3 |
| chr10_+_84591919 | 1.75 |
ENSMUST00000060397.13
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr5_-_146157711 | 1.71 |
ENSMUST00000169407.9
ENSMUST00000161331.8 ENSMUST00000159074.3 ENSMUST00000067837.10 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr19_+_24853039 | 1.70 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr19_-_20931566 | 1.67 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chr1_-_93406515 | 1.66 |
ENSMUST00000170883.8
ENSMUST00000189025.7 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr16_-_50252703 | 1.65 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr1_-_14374794 | 1.65 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_-_14374842 | 1.60 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_+_20742115 | 1.50 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr10_+_53213763 | 1.47 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr14_-_51134930 | 1.41 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr11_-_99213769 | 1.40 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr11_-_74228504 | 1.39 |
ENSMUST00000213831.2
|
Olfr410
|
olfactory receptor 410 |
| chr12_-_83609217 | 1.35 |
ENSMUST00000222448.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr2_+_70393782 | 1.35 |
ENSMUST00000123330.3
|
Gad1
|
glutamate decarboxylase 1 |
| chr19_-_43879031 | 1.35 |
ENSMUST00000212048.2
|
Dnmbp
|
dynamin binding protein |
| chr6_+_54017063 | 1.34 |
ENSMUST00000127323.3
|
Chn2
|
chimerin 2 |
| chr4_+_11579648 | 1.32 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr19_+_11943265 | 1.32 |
ENSMUST00000025590.11
|
Osbp
|
oxysterol binding protein |
| chr7_-_84339156 | 1.31 |
ENSMUST00000209117.2
ENSMUST00000207975.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr14_-_80008745 | 1.28 |
ENSMUST00000039568.11
ENSMUST00000195355.2 |
Pcdh8
|
protocadherin 8 |
| chr14_+_47069667 | 1.28 |
ENSMUST00000140114.3
ENSMUST00000133989.8 |
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr2_-_57003064 | 1.25 |
ENSMUST00000112627.2
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr10_+_63293284 | 1.24 |
ENSMUST00000105440.8
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr16_-_35891739 | 1.24 |
ENSMUST00000231351.2
ENSMUST00000004057.9 |
Fam162a
|
family with sequence similarity 162, member A |
| chr14_-_51134906 | 1.20 |
ENSMUST00000170855.2
|
Klhl33
|
kelch-like 33 |
| chr2_+_83786742 | 1.19 |
ENSMUST00000178325.2
|
Gm13698
|
predicted gene 13698 |
| chr14_-_30213408 | 1.18 |
ENSMUST00000112250.6
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr9_+_50466127 | 1.15 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr6_+_29468067 | 1.15 |
ENSMUST00000143101.4
ENSMUST00000149646.3 |
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr2_-_77349909 | 1.14 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr8_-_85663976 | 1.13 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr16_-_22676264 | 1.13 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr8_-_45747883 | 1.12 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chr16_+_35891991 | 1.11 |
ENSMUST00000164916.9
|
Ccdc58
|
coiled-coil domain containing 58 |
| chr19_+_8817883 | 1.10 |
ENSMUST00000086058.13
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr17_-_43978023 | 1.10 |
ENSMUST00000233442.2
|
Slc25a27
|
solute carrier family 25, member 27 |
| chr11_-_74228857 | 1.04 |
ENSMUST00000214490.2
|
Olfr410
|
olfactory receptor 410 |
| chr18_-_79152504 | 1.03 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr13_+_38388904 | 1.03 |
ENSMUST00000091641.13
ENSMUST00000178564.2 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr4_+_101407608 | 1.00 |
ENSMUST00000094953.11
ENSMUST00000106933.2 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr10_+_127256192 | 1.00 |
ENSMUST00000171434.8
|
R3hdm2
|
R3H domain containing 2 |
| chr1_-_189902868 | 0.99 |
ENSMUST00000177288.4
ENSMUST00000175916.8 |
Prox1
|
prospero homeobox 1 |
| chr3_-_32546380 | 0.97 |
ENSMUST00000164954.3
|
Kcnmb3
|
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| chr2_+_177969456 | 0.96 |
ENSMUST00000133267.3
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr8_-_68427217 | 0.96 |
ENSMUST00000098696.10
ENSMUST00000038959.16 ENSMUST00000093469.11 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr14_-_36820304 | 0.92 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr13_-_103042294 | 0.91 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr5_+_115373895 | 0.89 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr11_+_21522221 | 0.89 |
ENSMUST00000131135.3
ENSMUST00000020568.10 |
Wdpcp
|
WD repeat containing planar cell polarity effector |
| chr17_-_43978056 | 0.88 |
ENSMUST00000024705.6
|
Slc25a27
|
solute carrier family 25, member 27 |
| chr6_+_77219698 | 0.87 |
ENSMUST00000161677.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr17_+_93506590 | 0.87 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr2_-_112089627 | 0.87 |
ENSMUST00000043970.2
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr6_-_69521891 | 0.85 |
ENSMUST00000103356.4
|
Igkv4-57-1
|
immunoglobulin kappa variable 4-57-1 |
| chr2_+_111607774 | 0.85 |
ENSMUST00000214708.2
ENSMUST00000215244.2 |
Olfr1302
|
olfactory receptor 1302 |
| chrX_-_104918911 | 0.85 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr6_+_29467717 | 0.82 |
ENSMUST00000004396.13
|
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr10_+_4561974 | 0.82 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr18_+_37440497 | 0.82 |
ENSMUST00000056712.4
|
Pcdhb4
|
protocadherin beta 4 |
| chr6_+_77219627 | 0.81 |
ENSMUST00000159616.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr1_-_163141230 | 0.81 |
ENSMUST00000174397.3
ENSMUST00000075805.13 |
Prrx1
|
paired related homeobox 1 |
| chr17_-_46044768 | 0.81 |
ENSMUST00000178179.2
|
1600014C23Rik
|
RIKEN cDNA 1600014C23 gene |
| chr1_-_67077906 | 0.80 |
ENSMUST00000119559.8
ENSMUST00000149996.2 ENSMUST00000027149.12 ENSMUST00000113979.10 |
Lancl1
|
LanC (bacterial lantibiotic synthetase component C)-like 1 |
| chr18_-_20247666 | 0.79 |
ENSMUST00000224432.2
|
Dsc1
|
desmocollin 1 |
| chr2_-_119377675 | 0.79 |
ENSMUST00000133668.2
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr6_+_49372463 | 0.79 |
ENSMUST00000024171.14
ENSMUST00000163954.8 ENSMUST00000172459.8 |
Stk31
|
serine threonine kinase 31 |
| chr7_-_136915602 | 0.78 |
ENSMUST00000210774.2
|
Ebf3
|
early B cell factor 3 |
| chr18_+_86729645 | 0.78 |
ENSMUST00000122464.8
|
Cbln2
|
cerebellin 2 precursor protein |
| chr7_-_119744509 | 0.78 |
ENSMUST00000208874.2
ENSMUST00000033207.6 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr18_+_61096597 | 0.77 |
ENSMUST00000115295.9
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr18_-_61147272 | 0.77 |
ENSMUST00000025520.10
|
Slc6a7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 |
| chr13_-_103042554 | 0.76 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr13_+_83672654 | 0.76 |
ENSMUST00000199019.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chrX_-_94521712 | 0.74 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr4_+_150322151 | 0.74 |
ENSMUST00000141931.2
|
Eno1
|
enolase 1, alpha non-neuron |
| chr4_+_116544509 | 0.74 |
ENSMUST00000030454.6
|
Prdx1
|
peroxiredoxin 1 |
| chr7_+_141276575 | 0.74 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr5_+_76957812 | 0.73 |
ENSMUST00000120818.8
|
Cracd
|
capping protein inhibiting regulator of actin |
| chr3_+_41519289 | 0.71 |
ENSMUST00000168086.7
|
Jade1
|
jade family PHD finger 1 |
| chr17_+_93506435 | 0.71 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr1_-_163141278 | 0.70 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr12_+_38830812 | 0.70 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr6_+_8949669 | 0.70 |
ENSMUST00000060369.4
|
Nxph1
|
neurexophilin 1 |
| chr16_-_85698679 | 0.68 |
ENSMUST00000023611.7
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2) |
| chr1_-_92446141 | 0.67 |
ENSMUST00000189174.2
|
Olfr1414
|
olfactory receptor 1414 |
| chr12_-_84408576 | 0.67 |
ENSMUST00000021659.2
ENSMUST00000065536.9 |
Fam161b
|
family with sequence similarity 161, member B |
| chr8_+_46984016 | 0.67 |
ENSMUST00000152423.2
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr14_+_32321341 | 0.67 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr13_+_83672389 | 0.66 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr5_+_137059522 | 0.66 |
ENSMUST00000187382.2
|
Vgf
|
VGF nerve growth factor inducible |
| chr19_-_23425757 | 0.66 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chr18_+_37433852 | 0.66 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr11_+_97732108 | 0.65 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr13_+_58956077 | 0.65 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr9_-_37580478 | 0.65 |
ENSMUST00000011262.4
|
Panx3
|
pannexin 3 |
| chr1_+_42734051 | 0.64 |
ENSMUST00000239323.2
ENSMUST00000199521.5 ENSMUST00000176807.3 |
Pou3f3
Gm20646
|
POU domain, class 3, transcription factor 3 predicted gene 20646 |
| chr4_+_114678919 | 0.64 |
ENSMUST00000137570.2
|
Gm12830
|
predicted gene 12830 |
| chr11_-_33153587 | 0.63 |
ENSMUST00000037746.8
|
Tlx3
|
T cell leukemia, homeobox 3 |
| chr15_-_99717956 | 0.63 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr7_-_34755985 | 0.63 |
ENSMUST00000130491.3
|
Cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr19_-_38212544 | 0.62 |
ENSMUST00000067167.6
|
Fra10ac1
|
FRA10AC1 homolog (human) |
| chr5_+_139529643 | 0.62 |
ENSMUST00000174792.2
|
Uncx
|
UNC homeobox |
| chr7_-_121700958 | 0.61 |
ENSMUST00000139456.2
ENSMUST00000106471.9 ENSMUST00000123296.8 ENSMUST00000033157.10 |
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr3_-_122910662 | 0.61 |
ENSMUST00000051443.8
|
Synpo2
|
synaptopodin 2 |
| chr11_+_108814007 | 0.59 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chr1_-_165762469 | 0.59 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr8_-_68658694 | 0.59 |
ENSMUST00000212960.2
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr16_+_10630779 | 0.58 |
ENSMUST00000115822.2
|
Gm11172
|
predicted gene 11172 |
| chr18_+_37622518 | 0.57 |
ENSMUST00000055949.4
|
Pcdhb18
|
protocadherin beta 18 |
| chr5_+_115573055 | 0.57 |
ENSMUST00000136586.6
|
Msi1
|
musashi RNA-binding protein 1 |
| chr5_+_137059127 | 0.57 |
ENSMUST00000041543.9
ENSMUST00000186451.2 |
Vgf
|
VGF nerve growth factor inducible |
| chr6_-_39787813 | 0.57 |
ENSMUST00000114797.2
ENSMUST00000031978.9 |
Mrps33
|
mitochondrial ribosomal protein S33 |
| chr19_-_57185808 | 0.56 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_+_139753130 | 0.56 |
ENSMUST00000209314.3
ENSMUST00000213953.2 ENSMUST00000214272.2 ENSMUST00000215785.2 ENSMUST00000216023.2 |
Olfr523
|
olfactory receptor 523 |
| chr11_+_56902658 | 0.55 |
ENSMUST00000094179.11
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1) |
| chr18_-_32082624 | 0.54 |
ENSMUST00000064016.6
|
Gpr17
|
G protein-coupled receptor 17 |
| chr8_+_47439948 | 0.54 |
ENSMUST00000119686.2
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr2_+_79538124 | 0.54 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr3_+_93349637 | 0.54 |
ENSMUST00000064257.6
|
Tchh
|
trichohyalin |
| chr1_-_144124871 | 0.54 |
ENSMUST00000189061.7
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr1_+_93406809 | 0.52 |
ENSMUST00000112912.8
|
Septin2
|
septin 2 |
| chr12_+_56742413 | 0.52 |
ENSMUST00000001538.10
|
Pax9
|
paired box 9 |
| chr14_-_109151590 | 0.52 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr1_-_92446383 | 0.52 |
ENSMUST00000062353.12
|
Olfr1414
|
olfactory receptor 1414 |
| chr8_+_88978600 | 0.51 |
ENSMUST00000154115.2
|
Tent4b
|
terminal nucleotidyltransferase 4B |
| chr12_-_31763859 | 0.50 |
ENSMUST00000057783.6
ENSMUST00000236002.2 ENSMUST00000174480.3 ENSMUST00000176710.2 |
Gpr22
|
G protein-coupled receptor 22 |
| chr5_-_105491795 | 0.49 |
ENSMUST00000171587.2
|
Gbp11
|
guanylate binding protein 11 |
| chr10_-_63926044 | 0.49 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chrX_+_65696608 | 0.49 |
ENSMUST00000036043.5
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr11_+_56902624 | 0.48 |
ENSMUST00000036315.16
|
Gria1
|
glutamate receptor, ionotropic, AMPA1 (alpha 1) |
| chr19_-_57185988 | 0.48 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr1_-_144124787 | 0.48 |
ENSMUST00000185714.2
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr13_+_112454981 | 0.47 |
ENSMUST00000223871.2
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr2_-_115896279 | 0.47 |
ENSMUST00000110907.8
ENSMUST00000110908.9 |
Meis2
|
Meis homeobox 2 |
| chr17_-_84495364 | 0.46 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr14_-_54648057 | 0.46 |
ENSMUST00000200545.2
ENSMUST00000227967.2 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr1_-_5140504 | 0.45 |
ENSMUST00000147158.2
ENSMUST00000118000.8 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr1_+_17797257 | 0.45 |
ENSMUST00000159958.8
ENSMUST00000160305.8 ENSMUST00000095075.5 |
Crispld1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr13_+_58955506 | 0.45 |
ENSMUST00000079828.7
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr13_+_22656093 | 0.45 |
ENSMUST00000226330.2
ENSMUST00000226965.2 |
Vmn1r201
|
vomeronasal 1 receptor 201 |
| chr6_-_138404076 | 0.44 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chr9_+_7347369 | 0.43 |
ENSMUST00000005950.12
ENSMUST00000065079.6 |
Mmp12
|
matrix metallopeptidase 12 |
| chr7_+_131568167 | 0.43 |
ENSMUST00000045840.5
|
Gpr26
|
G protein-coupled receptor 26 |
| chr18_+_61096660 | 0.43 |
ENSMUST00000039904.7
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr5_-_84565218 | 0.42 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr13_+_58955675 | 0.42 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr10_+_34265969 | 0.41 |
ENSMUST00000105511.2
|
Col10a1
|
collagen, type X, alpha 1 |
| chr7_+_144391786 | 0.41 |
ENSMUST00000155320.8
|
Fgf3
|
fibroblast growth factor 3 |
| chr3_+_94744844 | 0.41 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr12_+_38830283 | 0.40 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chr16_-_76170714 | 0.40 |
ENSMUST00000231585.2
ENSMUST00000121927.8 |
Nrip1
|
nuclear receptor interacting protein 1 |
| chr6_-_52190299 | 0.40 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308 |
| chr12_+_38830081 | 0.40 |
ENSMUST00000095767.11
|
Etv1
|
ets variant 1 |
| chr18_+_65021974 | 0.39 |
ENSMUST00000225261.3
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 2.8 | 8.5 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.5 | 13.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 1.2 | 3.7 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.9 | 8.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.6 | 2.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.6 | 2.4 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.6 | 3.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.5 | 3.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 | 6.5 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.5 | 2.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.5 | 11.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.5 | 4.6 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 | 1.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.4 | 2.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.4 | 2.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 3.9 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.3 | 1.6 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.3 | 3.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 2.9 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 1.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.3 | 1.2 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.3 | 1.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 1.5 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.2 | 1.7 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 1.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 1.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 1.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 3.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 1.2 | GO:0097394 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.2 | 0.8 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 1.8 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.2 | 1.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.6 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 1.5 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 1.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 3.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.8 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 4.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 0.8 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.9 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.6 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 1.4 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 1.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.7 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.6 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.3 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 1.2 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 1.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 2.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 1.2 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 1.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.2 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.2 | GO:1902871 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 3.7 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 0.7 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 0.5 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 13.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.4 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.1 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.6 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 2.0 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.1 | 1.3 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:1902739 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 1.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 3.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.7 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.6 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 4.9 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 2.6 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 1.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.0 | 0.7 | GO:0071450 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.3 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.6 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 1.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.4 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.6 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 3.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 2.5 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.3 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 2.2 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.0 | 10.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 1.2 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.3 | 2.7 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 2.4 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.3 | 4.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 1.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 1.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 1.2 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.2 | 3.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 2.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 5.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.8 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 3.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 1.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.3 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 2.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 3.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 2.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 3.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.8 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 4.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.9 | 3.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.8 | 2.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.7 | 3.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.6 | 4.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.4 | 3.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.4 | 14.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.4 | 2.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 1.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.3 | 12.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 0.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 0.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 1.5 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 1.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.6 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.2 | 12.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 1.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.7 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 1.0 | GO:0099583 | AMPA glutamate receptor activity(GO:0004971) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 3.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 4.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 3.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 4.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 3.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 10.1 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 1.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 4.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 10.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.7 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 3.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0005234 | ionotropic glutamate receptor activity(GO:0004970) extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 2.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 2.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 10.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 5.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 3.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 3.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 10.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 3.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 3.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 8.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.6 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |