Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
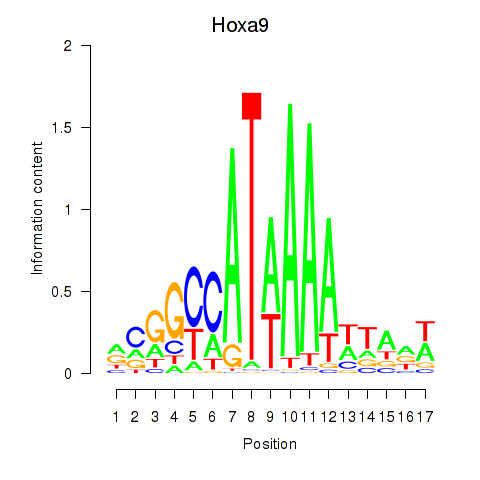
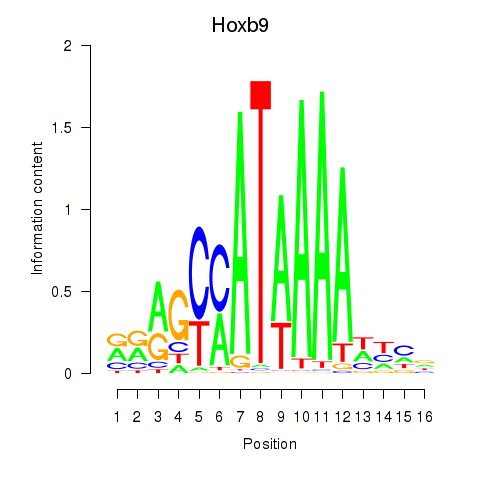
Results for Hoxa9_Hoxb9
Z-value: 0.55
Transcription factors associated with Hoxa9_Hoxb9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa9
|
ENSMUSG00000038227.16 | homeobox A9 |
|
Hoxb9
|
ENSMUSG00000020875.10 | homeobox B9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa9 | mm39_v1_chr6_-_52203146_52203176 | 0.25 | 1.4e-01 | Click! |
| Hoxb9 | mm39_v1_chr11_+_96162283_96162349 | 0.22 | 1.9e-01 | Click! |
Activity profile of Hoxa9_Hoxb9 motif
Sorted Z-values of Hoxa9_Hoxb9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142215595 | 3.98 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr1_-_45542442 | 1.32 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr17_-_43813664 | 1.31 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr18_+_20380397 | 0.95 |
ENSMUST00000054128.7
|
Dsg1c
|
desmoglein 1 gamma |
| chr10_+_128745214 | 0.68 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr2_-_151586063 | 0.67 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr2_+_163348728 | 0.53 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr3_+_98187743 | 0.52 |
ENSMUST00000120541.8
ENSMUST00000090746.3 |
Hmgcs2
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
| chr8_-_4155758 | 0.46 |
ENSMUST00000138439.2
ENSMUST00000145007.8 |
Cd209f
|
CD209f antigen |
| chr2_+_58645189 | 0.45 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr19_-_7319157 | 0.42 |
ENSMUST00000164205.8
ENSMUST00000165286.8 ENSMUST00000168324.2 ENSMUST00000032557.15 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr2_+_58644922 | 0.39 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr14_-_70561231 | 0.39 |
ENSMUST00000151011.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr19_-_56536646 | 0.38 |
ENSMUST00000182276.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr6_+_29694181 | 0.36 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr2_+_80145805 | 0.36 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr4_-_136329953 | 0.36 |
ENSMUST00000105847.8
ENSMUST00000116273.9 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chrX_-_133442596 | 0.34 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr17_+_36172210 | 0.33 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr5_+_145141333 | 0.33 |
ENSMUST00000085671.10
ENSMUST00000031601.8 |
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr8_+_4184652 | 0.33 |
ENSMUST00000130372.3
|
Cd209g
|
CD209g antigen |
| chr3_+_57332735 | 0.32 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr15_+_6552270 | 0.31 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr15_-_51855073 | 0.31 |
ENSMUST00000022927.11
|
Rad21
|
RAD21 cohesin complex component |
| chr9_-_56151334 | 0.29 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr6_-_87958611 | 0.29 |
ENSMUST00000056403.7
|
H1f10
|
H1.10 linker histone |
| chr2_-_27354006 | 0.28 |
ENSMUST00000164296.8
|
Brd3
|
bromodomain containing 3 |
| chr15_+_81820954 | 0.27 |
ENSMUST00000038757.8
ENSMUST00000230633.2 |
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr3_+_66892979 | 0.27 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr6_-_139478919 | 0.26 |
ENSMUST00000170650.3
|
Rergl
|
RERG/RAS-like |
| chr9_-_63929308 | 0.26 |
ENSMUST00000179458.2
ENSMUST00000041029.6 |
Smad6
|
SMAD family member 6 |
| chr2_-_51039112 | 0.26 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr2_-_80277969 | 0.25 |
ENSMUST00000028389.4
|
Frzb
|
frizzled-related protein |
| chr19_+_56536685 | 0.25 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr4_+_132857816 | 0.23 |
ENSMUST00000084241.12
ENSMUST00000138831.2 |
Wasf2
|
WASP family, member 2 |
| chr1_+_88139678 | 0.23 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr1_-_125362321 | 0.22 |
ENSMUST00000191544.7
|
Actr3
|
ARP3 actin-related protein 3 |
| chr14_-_31503869 | 0.22 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr9_-_107648144 | 0.22 |
ENSMUST00000183248.3
ENSMUST00000182022.8 ENSMUST00000035199.13 ENSMUST00000182659.8 |
Rbm5
|
RNA binding motif protein 5 |
| chr2_+_29509704 | 0.21 |
ENSMUST00000095087.11
ENSMUST00000091146.12 ENSMUST00000102872.11 ENSMUST00000147755.10 |
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr7_+_27977141 | 0.20 |
ENSMUST00000094651.4
|
Eid2b
|
EP300 interacting inhibitor of differentiation 2B |
| chr7_-_101519973 | 0.20 |
ENSMUST00000126204.8
ENSMUST00000155311.2 ENSMUST00000106983.8 ENSMUST00000123630.8 |
Folr1
|
folate receptor 1 (adult) |
| chr17_+_36172235 | 0.20 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr17_-_71305003 | 0.20 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr19_-_56536443 | 0.20 |
ENSMUST00000182059.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chrX_+_55493325 | 0.19 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr10_+_3822667 | 0.19 |
ENSMUST00000136671.8
ENSMUST00000042438.13 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr17_-_35081129 | 0.19 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr2_+_84818538 | 0.19 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr15_+_3300249 | 0.19 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr5_+_137786031 | 0.18 |
ENSMUST00000035852.14
|
Zcwpw1
|
zinc finger, CW type with PWWP domain 1 |
| chr11_-_83421333 | 0.17 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr17_-_35081456 | 0.17 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr3_+_66893280 | 0.17 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr3_+_66893031 | 0.17 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr1_+_107439145 | 0.17 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr1_-_97689263 | 0.16 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr11_-_99134885 | 0.16 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr4_-_49597837 | 0.16 |
ENSMUST00000042750.3
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr13_-_101829070 | 0.16 |
ENSMUST00000187009.7
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr7_-_101513300 | 0.15 |
ENSMUST00000106981.8
|
Folr1
|
folate receptor 1 (adult) |
| chr19_-_7319211 | 0.15 |
ENSMUST00000171393.8
|
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr3_+_108561247 | 0.14 |
ENSMUST00000124384.8
ENSMUST00000029483.15 |
Clcc1
|
chloride channel CLIC-like 1 |
| chr7_-_101519914 | 0.14 |
ENSMUST00000106985.8
ENSMUST00000151706.8 |
Folr1
|
folate receptor 1 (adult) |
| chr17_+_14087827 | 0.14 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr3_+_82933383 | 0.13 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr3_+_32490525 | 0.13 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr5_-_73349191 | 0.13 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr14_+_20724378 | 0.12 |
ENSMUST00000224492.2
ENSMUST00000223751.2 ENSMUST00000225108.2 ENSMUST00000224754.2 |
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr19_-_33764859 | 0.12 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr6_-_122317484 | 0.11 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr4_-_135780660 | 0.11 |
ENSMUST00000102536.11
|
Rpl11
|
ribosomal protein L11 |
| chr17_+_33857030 | 0.11 |
ENSMUST00000052079.8
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1 |
| chr17_-_29226700 | 0.11 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38 |
| chr9_+_103917821 | 0.10 |
ENSMUST00000216593.2
ENSMUST00000147249.3 |
Nphp3
Gm28305
|
nephronophthisis 3 (adolescent) predicted gene 28305 |
| chr18_+_37898633 | 0.10 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr6_-_141719536 | 0.10 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr1_-_128030148 | 0.10 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr6_+_108771840 | 0.10 |
ENSMUST00000204483.2
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr3_+_108561223 | 0.10 |
ENSMUST00000106609.8
|
Clcc1
|
chloride channel CLIC-like 1 |
| chr7_-_101519872 | 0.09 |
ENSMUST00000106986.9
|
Folr1
|
folate receptor 1 (adult) |
| chrX_+_158623460 | 0.09 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr17_+_87270504 | 0.09 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr2_-_109111064 | 0.09 |
ENSMUST00000147770.2
|
Mettl15
|
methyltransferase like 15 |
| chr9_+_19620729 | 0.09 |
ENSMUST00000217450.4
ENSMUST00000212013.4 |
Olfr857
|
olfactory receptor 857 |
| chr7_-_80055168 | 0.09 |
ENSMUST00000107362.10
ENSMUST00000135306.3 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr9_+_121196034 | 0.09 |
ENSMUST00000210636.2
ENSMUST00000045903.8 |
Trak1
|
trafficking protein, kinesin binding 1 |
| chr1_-_156546600 | 0.09 |
ENSMUST00000122424.8
ENSMUST00000086153.8 |
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr11_+_66969119 | 0.09 |
ENSMUST00000108689.8
ENSMUST00000007301.14 ENSMUST00000165221.2 |
Myh3
|
myosin, heavy polypeptide 3, skeletal muscle, embryonic |
| chr4_+_44012638 | 0.08 |
ENSMUST00000107847.10
ENSMUST00000170241.8 ENSMUST00000107849.10 ENSMUST00000107851.10 ENSMUST00000107845.4 |
Clta
|
clathrin, light polypeptide (Lca) |
| chr3_+_108561294 | 0.08 |
ENSMUST00000106613.2
|
Clcc1
|
chloride channel CLIC-like 1 |
| chr14_+_20724366 | 0.08 |
ENSMUST00000048657.10
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr9_+_7571397 | 0.08 |
ENSMUST00000120900.8
ENSMUST00000151853.8 ENSMUST00000152878.3 |
Mmp27
|
matrix metallopeptidase 27 |
| chr7_-_135130374 | 0.08 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr6_-_56681657 | 0.08 |
ENSMUST00000176595.3
ENSMUST00000170382.5 ENSMUST00000203958.2 |
Lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr19_-_57185988 | 0.08 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr3_-_113367891 | 0.07 |
ENSMUST00000142505.9
|
Amy1
|
amylase 1, salivary |
| chr16_-_19241884 | 0.07 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr7_+_142086880 | 0.07 |
ENSMUST00000210803.2
|
Mrpl23
|
mitochondrial ribosomal protein L23 |
| chr13_-_59890564 | 0.07 |
ENSMUST00000055343.3
|
1700014D04Rik
|
RIKEN cDNA 1700014D04 gene |
| chr5_+_31609772 | 0.07 |
ENSMUST00000202061.4
ENSMUST00000076264.8 ENSMUST00000201450.4 |
Zfp512
|
zinc finger protein 512 |
| chr7_+_142086749 | 0.07 |
ENSMUST00000038675.7
|
Mrpl23
|
mitochondrial ribosomal protein L23 |
| chr4_-_49549489 | 0.07 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr9_-_101076198 | 0.06 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr8_-_34333573 | 0.06 |
ENSMUST00000183062.2
|
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr4_+_145311759 | 0.06 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268 |
| chr6_+_67838100 | 0.06 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr8_+_10027707 | 0.06 |
ENSMUST00000139793.8
ENSMUST00000048216.6 |
Abhd13
|
abhydrolase domain containing 13 |
| chr13_-_111686445 | 0.06 |
ENSMUST00000132880.3
ENSMUST00000231096.2 |
Gm15290
Gm49496
|
predicted gene 15290 predicted gene, 49496 |
| chrX_+_158086253 | 0.06 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr17_+_37274714 | 0.06 |
ENSMUST00000209623.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr2_-_45002902 | 0.05 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_31609802 | 0.05 |
ENSMUST00000202244.4
|
Zfp512
|
zinc finger protein 512 |
| chr19_-_57185928 | 0.05 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr2_+_85804239 | 0.05 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr11_+_67061837 | 0.05 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr7_+_44711853 | 0.05 |
ENSMUST00000107829.9
ENSMUST00000003513.11 ENSMUST00000211465.2 ENSMUST00000210088.2 ENSMUST00000210520.2 |
Nosip
|
nitric oxide synthase interacting protein |
| chr10_+_17672004 | 0.05 |
ENSMUST00000037964.7
|
Txlnb
|
taxilin beta |
| chr11_+_67061908 | 0.05 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr7_+_110367375 | 0.05 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr6_+_134958681 | 0.05 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr4_+_115594918 | 0.05 |
ENSMUST00000106525.9
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr15_-_78947038 | 0.04 |
ENSMUST00000151889.8
ENSMUST00000040676.11 |
Ankrd54
|
ankyrin repeat domain 54 |
| chr2_+_85809620 | 0.04 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr5_+_63969706 | 0.04 |
ENSMUST00000081747.8
ENSMUST00000196575.5 |
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chrX_+_110920704 | 0.04 |
ENSMUST00000131304.2
|
Tex16
|
testis expressed gene 16 |
| chr7_-_19967753 | 0.04 |
ENSMUST00000168580.2
|
Vmn1r242
|
vomeronasal 1 receptor 242 |
| chr4_+_115594951 | 0.04 |
ENSMUST00000106522.9
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr2_-_88768449 | 0.04 |
ENSMUST00000215205.2
ENSMUST00000213412.2 |
Olfr1211
|
olfactory receptor 1211 |
| chr6_-_70217956 | 0.04 |
ENSMUST00000196275.2
|
Gm43218
|
predicted gene 43218 |
| chr5_+_143864993 | 0.04 |
ENSMUST00000172367.3
|
Gm42421
|
predicted gene, 42421 |
| chr16_+_18247666 | 0.04 |
ENSMUST00000144233.3
|
Txnrd2
|
thioredoxin reductase 2 |
| chr10_+_25308466 | 0.04 |
ENSMUST00000219224.2
ENSMUST00000219166.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr6_+_51500881 | 0.04 |
ENSMUST00000049152.15
|
Snx10
|
sorting nexin 10 |
| chr4_-_58787716 | 0.04 |
ENSMUST00000216719.2
|
Olfr267
|
olfactory receptor 267 |
| chr16_+_26281885 | 0.04 |
ENSMUST00000161053.8
ENSMUST00000115302.2 |
Cldn16
|
claudin 16 |
| chr1_-_171854818 | 0.04 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr9_-_60594742 | 0.04 |
ENSMUST00000114032.8
ENSMUST00000166168.8 ENSMUST00000132366.2 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr12_+_71184614 | 0.03 |
ENSMUST00000045907.16
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr4_+_146586445 | 0.03 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr6_-_24168082 | 0.03 |
ENSMUST00000031713.9
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chr7_+_55699883 | 0.03 |
ENSMUST00000205653.2
ENSMUST00000076226.13 |
Herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr6_+_70675416 | 0.03 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr9_+_38259707 | 0.03 |
ENSMUST00000217063.2
|
Olfr898
|
olfactory receptor 898 |
| chr8_+_121215155 | 0.03 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr19_+_13628455 | 0.03 |
ENSMUST00000213900.2
|
Olfr1490
|
olfactory receptor 1490 |
| chr1_+_59296065 | 0.03 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr11_+_115294560 | 0.03 |
ENSMUST00000153983.8
ENSMUST00000106539.10 ENSMUST00000103036.5 |
Mrpl58
|
mitochondrial ribosomal protein L58 |
| chr13_-_48746836 | 0.03 |
ENSMUST00000238995.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr19_+_13208692 | 0.03 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr18_-_31450095 | 0.03 |
ENSMUST00000139924.2
ENSMUST00000153060.8 |
Rit2
|
Ras-like without CAAX 2 |
| chr2_+_89821565 | 0.03 |
ENSMUST00000111509.3
ENSMUST00000213909.3 |
Olfr1261
|
olfactory receptor 1261 |
| chr9_+_111014254 | 0.02 |
ENSMUST00000198986.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr16_-_19132814 | 0.02 |
ENSMUST00000216157.2
|
Olfr164
|
olfactory receptor 164 |
| chrY_-_9132561 | 0.02 |
ENSMUST00000171947.3
|
Gm21292
|
predicted gene, 21292 |
| chr1_+_66403774 | 0.02 |
ENSMUST00000077355.12
ENSMUST00000114012.8 |
Map2
|
microtubule-associated protein 2 |
| chr2_-_32977182 | 0.02 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chrY_+_2904133 | 0.02 |
ENSMUST00000166474.3
|
Gm10352
|
predicted gene 10352 |
| chr11_-_99996452 | 0.02 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr11_-_99941369 | 0.02 |
ENSMUST00000007318.2
|
Krt31
|
keratin 31 |
| chr6_-_70364222 | 0.02 |
ENSMUST00000103392.3
ENSMUST00000195945.2 |
Igkv8-16
|
immunoglobulin kappa variable 8-16 |
| chrX_+_110801086 | 0.02 |
ENSMUST00000207962.2
|
Gm45194
|
predicted gene 45194 |
| chr9_-_101128976 | 0.02 |
ENSMUST00000075941.12
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr2_-_181240921 | 0.02 |
ENSMUST00000060173.9
|
Samd10
|
sterile alpha motif domain containing 10 |
| chr10_-_130265572 | 0.02 |
ENSMUST00000171811.4
|
Vmn2r85
|
vomeronasal 2, receptor 85 |
| chr16_+_88574717 | 0.01 |
ENSMUST00000009191.4
ENSMUST00000238221.2 |
Gm5965
|
predicted gene 5965 |
| chr6_+_146944253 | 0.01 |
ENSMUST00000036111.10
|
Mrps35
|
mitochondrial ribosomal protein S35 |
| chr3_+_108498595 | 0.01 |
ENSMUST00000051145.15
|
Wdr47
|
WD repeat domain 47 |
| chr10_+_129153986 | 0.01 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr10_+_129306867 | 0.01 |
ENSMUST00000213222.2
|
Olfr788
|
olfactory receptor 788 |
| chr17_-_29226965 | 0.01 |
ENSMUST00000009138.13
ENSMUST00000119274.3 |
Stk38
|
serine/threonine kinase 38 |
| chr3_-_95014218 | 0.01 |
ENSMUST00000107233.9
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr16_-_22630327 | 0.01 |
ENSMUST00000040592.6
|
Crygs
|
crystallin, gamma S |
| chr7_-_101519791 | 0.01 |
ENSMUST00000124026.8
|
Folr1
|
folate receptor 1 (adult) |
| chr3_+_84859453 | 0.01 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr6_-_122317457 | 0.01 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chr6_+_70565173 | 0.01 |
ENSMUST00000103398.2
|
Igkv3-9
|
immunoglobulin kappa variable 3-9 |
| chr2_+_89821818 | 0.01 |
ENSMUST00000216953.3
|
Olfr1261
|
olfactory receptor 1261 |
| chr15_+_81821112 | 0.01 |
ENSMUST00000135663.2
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr6_+_70648743 | 0.00 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr2_+_36674288 | 0.00 |
ENSMUST00000213498.2
ENSMUST00000214909.3 ENSMUST00000215199.2 |
Olfr348
|
olfactory receptor 348 |
| chr3_+_137770813 | 0.00 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr13_+_83723743 | 0.00 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr19_-_13075176 | 0.00 |
ENSMUST00000208913.2
ENSMUST00000215229.2 |
Olfr1457
|
olfactory receptor 1457 |
| chr7_-_20646272 | 0.00 |
ENSMUST00000172322.2
|
Vmn1r118
|
vomeronasal 1 receptor 118 |
| chr7_-_20437141 | 0.00 |
ENSMUST00000179511.2
|
Vmn1r111
|
vomeronasal 1 receptor 111 |
| chr7_-_21280015 | 0.00 |
ENSMUST00000179390.2
|
Vmn1r131
|
vomeronasal 1 receptor 131 |
| chr7_-_21728322 | 0.00 |
ENSMUST00000179206.2
|
Vmn1r138
|
vomeronasal 1 receptor 138 |
| chr7_-_22391742 | 0.00 |
ENSMUST00000178871.2
|
Vmn1r257
|
vomeronasal 1 receptor 257 |
| chr7_-_22951107 | 0.00 |
ENSMUST00000164045.2
|
Vmn1r166
|
vomeronasal 1 receptor 166 |
| chr7_-_21928909 | 0.00 |
ENSMUST00000167253.2
|
Vmn1r252
|
vomeronasal 1 receptor 252 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa9_Hoxb9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.3 | 1.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.5 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.1 | 0.7 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 0.6 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.6 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.2 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.5 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.7 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.3 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.0 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.2 | 0.5 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.8 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 4.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.2 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.6 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |