Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
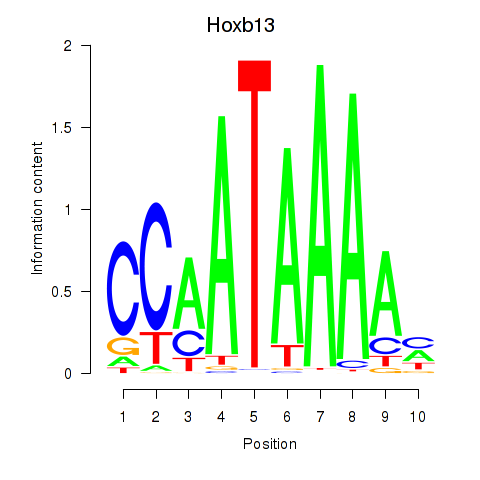
Results for Hoxb13
Z-value: 1.38
Transcription factors associated with Hoxb13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb13
|
ENSMUSG00000049604.4 | homeobox B13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb13 | mm39_v1_chr11_+_96085118_96085142 | -0.08 | 6.5e-01 | Click! |
Activity profile of Hoxb13 motif
Sorted Z-values of Hoxb13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_61592331 | 19.90 |
ENSMUST00000098040.4
|
Mup18
|
major urinary protein 18 |
| chr4_-_61259997 | 19.04 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr4_-_61259801 | 17.64 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr19_+_40078132 | 17.38 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr4_-_60538151 | 16.91 |
ENSMUST00000098047.3
|
Mup10
|
major urinary protein 10 |
| chr4_-_60377932 | 16.64 |
ENSMUST00000107506.9
ENSMUST00000122381.8 ENSMUST00000118759.8 ENSMUST00000132829.3 |
Mup9
|
major urinary protein 9 |
| chr4_-_60618357 | 16.55 |
ENSMUST00000084544.5
ENSMUST00000098046.10 |
Mup11
|
major urinary protein 11 |
| chr8_-_25066313 | 16.06 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr4_-_60139857 | 15.61 |
ENSMUST00000107490.5
ENSMUST00000074700.9 |
Mup2
|
major urinary protein 2 |
| chr4_-_60070411 | 15.53 |
ENSMUST00000079697.10
ENSMUST00000125282.2 ENSMUST00000166098.8 |
Mup7
|
major urinary protein 7 |
| chr4_-_61184270 | 15.49 |
ENSMUST00000072678.6
ENSMUST00000098042.10 |
Mup13
|
major urinary protein 13 |
| chr4_-_60777462 | 15.34 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr4_-_61437704 | 14.97 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr4_-_61357980 | 14.65 |
ENSMUST00000095049.5
|
Mup15
|
major urinary protein 15 |
| chr4_-_61514108 | 13.30 |
ENSMUST00000107484.2
|
Mup17
|
major urinary protein 17 |
| chr4_-_60222580 | 13.08 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr19_-_8382424 | 13.05 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr4_-_60697274 | 11.79 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr4_-_61972348 | 11.13 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr4_-_60457902 | 11.12 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr19_+_39275518 | 10.55 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr6_+_121983720 | 9.68 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chr5_-_145946408 | 8.00 |
ENSMUST00000138870.2
ENSMUST00000068317.13 |
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25 |
| chr7_+_43856724 | 7.75 |
ENSMUST00000077354.5
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr19_-_8109346 | 7.35 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr8_-_94006345 | 7.32 |
ENSMUST00000034178.9
|
Ces1f
|
carboxylesterase 1F |
| chr6_-_85846110 | 7.30 |
ENSMUST00000045008.8
|
Nat8f2
|
N-acetyltransferase 8 (GCN5-related) family member 2 |
| chr5_+_146016064 | 6.46 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr15_+_6474808 | 6.08 |
ENSMUST00000022749.17
ENSMUST00000239466.2 |
C9
|
complement component 9 |
| chr12_-_72675624 | 5.92 |
ENSMUST00000208039.2
ENSMUST00000207585.2 |
Gm4756
|
predicted gene 4756 |
| chr5_-_87485023 | 5.89 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr1_-_121260298 | 5.87 |
ENSMUST00000071064.13
|
Insig2
|
insulin induced gene 2 |
| chr6_+_42222841 | 5.69 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr15_-_77295234 | 5.38 |
ENSMUST00000089452.6
ENSMUST00000081776.11 |
Apol9a
|
apolipoprotein L 9a |
| chr19_+_12610870 | 5.33 |
ENSMUST00000119960.2
|
Glyat
|
glycine-N-acyltransferase |
| chr7_-_19432129 | 5.23 |
ENSMUST00000172808.2
ENSMUST00000174191.2 |
Apoe
|
apolipoprotein E |
| chrX_+_59044796 | 5.13 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr15_+_77613239 | 4.97 |
ENSMUST00000230979.2
ENSMUST00000109775.4 |
Apol9b
|
apolipoprotein L 9b |
| chr11_-_11848107 | 4.90 |
ENSMUST00000178704.8
|
Ddc
|
dopa decarboxylase |
| chr1_-_121260274 | 4.90 |
ENSMUST00000161068.2
|
Insig2
|
insulin induced gene 2 |
| chr19_+_12610668 | 4.68 |
ENSMUST00000044976.12
|
Glyat
|
glycine-N-acyltransferase |
| chr1_-_192946359 | 4.45 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr13_-_24098981 | 4.23 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr1_+_13738967 | 4.11 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr1_-_130589349 | 4.07 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr5_-_89583469 | 4.05 |
ENSMUST00000200534.2
|
Gc
|
vitamin D binding protein |
| chr1_-_130589321 | 4.00 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr5_-_87572060 | 3.96 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr1_+_74752710 | 3.94 |
ENSMUST00000027356.7
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr15_+_77613348 | 3.90 |
ENSMUST00000230742.2
|
Apol9b
|
apolipoprotein L 9b |
| chr13_-_24098951 | 3.88 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr14_+_66205932 | 3.78 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr2_-_86180622 | 3.74 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr19_+_38995463 | 3.70 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr7_+_107166925 | 3.69 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr8_-_93956143 | 3.69 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr9_+_78164402 | 3.52 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chr14_+_66207163 | 3.35 |
ENSMUST00000153460.8
|
Clu
|
clusterin |
| chrM_+_3906 | 3.31 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr14_-_52150804 | 3.14 |
ENSMUST00000004673.15
ENSMUST00000111632.5 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr4_-_96552349 | 3.14 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr12_-_103623418 | 3.11 |
ENSMUST00000044159.7
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr15_-_34495329 | 3.09 |
ENSMUST00000022946.6
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr3_-_146321341 | 2.97 |
ENSMUST00000200633.2
|
Dnase2b
|
deoxyribonuclease II beta |
| chr5_-_87288177 | 2.96 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr17_+_31652029 | 2.91 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr17_+_31652073 | 2.88 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr3_+_129630380 | 2.85 |
ENSMUST00000077918.7
|
Cfi
|
complement component factor i |
| chr19_-_39637489 | 2.78 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr14_-_47426863 | 2.76 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr5_+_9163244 | 2.76 |
ENSMUST00000198935.2
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr8_+_96078886 | 2.75 |
ENSMUST00000034243.7
|
Mmp15
|
matrix metallopeptidase 15 |
| chr5_+_115061293 | 2.75 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr16_-_10360893 | 2.71 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr16_+_91066602 | 2.70 |
ENSMUST00000056882.7
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr15_+_54975713 | 2.56 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr5_+_137979763 | 2.55 |
ENSMUST00000035390.7
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chr7_-_25239229 | 2.49 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr13_-_42001075 | 2.48 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr14_-_61794330 | 2.43 |
ENSMUST00000022497.15
|
Spryd7
|
SPRY domain containing 7 |
| chr10_-_115198093 | 2.41 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr2_+_121978156 | 2.40 |
ENSMUST00000102476.5
|
B2m
|
beta-2 microglobulin |
| chr6_+_124489364 | 2.32 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chr12_-_72675670 | 2.27 |
ENSMUST00000209038.2
|
Gm4756
|
predicted gene 4756 |
| chr8_-_70591746 | 2.27 |
ENSMUST00000212320.2
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr19_-_6117815 | 2.27 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr16_-_23807602 | 2.22 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr16_-_45975440 | 2.22 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr19_+_4771089 | 2.22 |
ENSMUST00000238976.3
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr19_-_7780025 | 2.18 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr5_+_114313940 | 2.18 |
ENSMUST00000146841.2
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr6_+_149043011 | 2.15 |
ENSMUST00000179873.8
ENSMUST00000047531.16 ENSMUST00000111548.8 ENSMUST00000111547.2 ENSMUST00000134306.8 ENSMUST00000147934.4 |
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr6_+_71176811 | 2.12 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr4_-_107975723 | 2.11 |
ENSMUST00000030340.15
|
Scp2
|
sterol carrier protein 2, liver |
| chr4_+_98919183 | 2.09 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr1_-_72251466 | 2.07 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr8_-_118398264 | 2.06 |
ENSMUST00000037955.14
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr4_+_43493344 | 2.06 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr7_+_132212349 | 2.05 |
ENSMUST00000033241.6
ENSMUST00000106170.8 |
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr13_-_42000958 | 2.05 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr8_-_122671588 | 2.04 |
ENSMUST00000057653.8
|
Car5a
|
carbonic anhydrase 5a, mitochondrial |
| chr1_+_88022776 | 2.04 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr13_-_42001102 | 2.04 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr6_+_72575458 | 2.02 |
ENSMUST00000070597.13
ENSMUST00000176364.8 ENSMUST00000176168.3 |
Retsat
|
retinol saturase (all trans retinol 13,14 reductase) |
| chr8_+_87350672 | 2.01 |
ENSMUST00000034141.18
ENSMUST00000122188.10 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr2_-_91025441 | 2.01 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr8_+_45522174 | 2.00 |
ENSMUST00000067984.9
|
Mtnr1a
|
melatonin receptor 1A |
| chr4_+_148225139 | 1.98 |
ENSMUST00000140049.8
ENSMUST00000105707.2 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr11_-_116080361 | 1.96 |
ENSMUST00000148601.2
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr13_-_4659120 | 1.96 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr1_-_180845054 | 1.95 |
ENSMUST00000036928.12
ENSMUST00000111068.8 |
Ephx1
|
epoxide hydrolase 1, microsomal |
| chr18_+_38551960 | 1.94 |
ENSMUST00000236085.2
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr8_-_70591799 | 1.93 |
ENSMUST00000075724.9
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr1_+_167135933 | 1.91 |
ENSMUST00000195015.6
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr3_+_89366632 | 1.90 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase |
| chr3_+_89366425 | 1.89 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr2_+_162829422 | 1.88 |
ENSMUST00000117123.2
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr2_+_162829250 | 1.88 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr10_-_115197775 | 1.88 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr6_-_3988900 | 1.86 |
ENSMUST00000183682.3
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr7_-_44711771 | 1.85 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr10_-_116732813 | 1.85 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr9_-_50466470 | 1.82 |
ENSMUST00000119103.2
|
Bco2
|
beta-carotene oxygenase 2 |
| chr19_+_24853039 | 1.81 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr11_+_96920751 | 1.80 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr15_-_31453708 | 1.80 |
ENSMUST00000110408.3
|
Ropn1l
|
ropporin 1-like |
| chr5_-_87686048 | 1.79 |
ENSMUST00000031199.11
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr6_-_23839136 | 1.79 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr19_+_30210320 | 1.78 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr11_-_73215442 | 1.78 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr15_-_77283286 | 1.78 |
ENSMUST00000175919.8
ENSMUST00000176074.9 |
Apol7a
|
apolipoprotein L 7a |
| chr19_+_39980868 | 1.76 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr12_-_103706774 | 1.75 |
ENSMUST00000186166.7
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr2_-_91025380 | 1.71 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_-_140590605 | 1.71 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr6_+_145561483 | 1.69 |
ENSMUST00000087445.7
|
Tuba3b
|
tubulin, alpha 3B |
| chr2_-_144392696 | 1.67 |
ENSMUST00000028915.6
|
Rbbp9
|
retinoblastoma binding protein 9, serine hydrolase |
| chr15_-_74869684 | 1.67 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr6_+_121709891 | 1.63 |
ENSMUST00000204124.2
|
Gm7298
|
predicted gene 7298 |
| chr18_+_38552011 | 1.63 |
ENSMUST00000025293.5
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr19_+_10160283 | 1.63 |
ENSMUST00000235160.2
|
Fads1
|
fatty acid desaturase 1 |
| chr3_-_96104886 | 1.63 |
ENSMUST00000016087.4
|
Bola1
|
bolA-like 1 (E. coli) |
| chr14_+_51333816 | 1.61 |
ENSMUST00000169895.3
|
Rnase4
|
ribonuclease, RNase A family 4 |
| chr2_-_32271833 | 1.60 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr16_+_10363222 | 1.60 |
ENSMUST00000155633.8
ENSMUST00000066345.15 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr1_-_161078723 | 1.60 |
ENSMUST00000051925.5
ENSMUST00000071718.12 |
Prdx6
|
peroxiredoxin 6 |
| chr7_+_127399776 | 1.58 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr13_-_86194889 | 1.57 |
ENSMUST00000131011.2
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr2_-_136733282 | 1.56 |
ENSMUST00000028730.13
ENSMUST00000110089.9 ENSMUST00000227806.2 |
Mkks
ENSMUSG00000115423.3
|
McKusick-Kaufman syndrome novel protein |
| chr7_+_119206233 | 1.56 |
ENSMUST00000126367.8
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr16_+_22926162 | 1.55 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr16_+_22926504 | 1.55 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr2_+_24944407 | 1.55 |
ENSMUST00000102931.11
ENSMUST00000074422.14 ENSMUST00000132172.8 ENSMUST00000114388.8 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr4_-_108075119 | 1.54 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr2_+_155907100 | 1.54 |
ENSMUST00000038860.12
|
Spag4
|
sperm associated antigen 4 |
| chr13_+_32985990 | 1.53 |
ENSMUST00000021832.7
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chr2_-_91025492 | 1.52 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr10_-_18110682 | 1.52 |
ENSMUST00000052648.9
ENSMUST00000080860.13 ENSMUST00000173243.8 |
Ccdc28a
|
coiled-coil domain containing 28A |
| chr3_-_89319088 | 1.52 |
ENSMUST00000107429.10
ENSMUST00000129308.9 ENSMUST00000107426.8 ENSMUST00000050398.11 ENSMUST00000162701.2 |
Flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr15_+_31224616 | 1.51 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr1_-_36283326 | 1.48 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr9_-_36708599 | 1.47 |
ENSMUST00000238932.2
ENSMUST00000115086.13 |
Ei24
|
etoposide induced 2.4 mRNA |
| chr12_-_104120105 | 1.47 |
ENSMUST00000085050.4
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr3_-_10396418 | 1.45 |
ENSMUST00000191670.6
ENSMUST00000065938.15 ENSMUST00000118410.8 |
Impa1
|
inositol (myo)-1(or 4)-monophosphatase 1 |
| chr13_+_4099001 | 1.45 |
ENSMUST00000118717.10
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr14_+_67470884 | 1.43 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr17_-_30831576 | 1.43 |
ENSMUST00000235171.2
ENSMUST00000236335.2 ENSMUST00000167624.2 |
Glo1
|
glyoxalase 1 |
| chr8_+_57004125 | 1.43 |
ENSMUST00000110322.9
ENSMUST00000040218.13 ENSMUST00000210863.2 |
Fbxo8
|
F-box protein 8 |
| chr19_+_10160249 | 1.43 |
ENSMUST00000010807.6
|
Fads1
|
fatty acid desaturase 1 |
| chr11_-_113600838 | 1.43 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr3_-_63836796 | 1.42 |
ENSMUST00000061706.7
|
E130311K13Rik
|
RIKEN cDNA E130311K13 gene |
| chr17_-_59320257 | 1.42 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr4_+_139350152 | 1.42 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr14_-_101878106 | 1.40 |
ENSMUST00000100339.9
|
Commd6
|
COMM domain containing 6 |
| chr2_+_34764408 | 1.39 |
ENSMUST00000113068.9
ENSMUST00000047447.13 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr9_-_51240201 | 1.39 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr7_+_29931309 | 1.38 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr18_-_38342815 | 1.37 |
ENSMUST00000057185.13
|
Pcdh1
|
protocadherin 1 |
| chr15_-_81244940 | 1.36 |
ENSMUST00000023040.9
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr11_+_94102255 | 1.35 |
ENSMUST00000041589.6
|
Tob1
|
transducer of ErbB-2.1 |
| chr11_+_105956867 | 1.33 |
ENSMUST00000002048.8
|
Taco1
|
translational activator of mitochondrially encoded cytochrome c oxidase I |
| chr2_-_91067212 | 1.32 |
ENSMUST00000111352.8
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr15_+_32920869 | 1.32 |
ENSMUST00000022871.7
|
Sdc2
|
syndecan 2 |
| chr8_-_25528972 | 1.31 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr9_-_44710480 | 1.31 |
ENSMUST00000214833.2
ENSMUST00000213972.2 ENSMUST00000214431.2 ENSMUST00000213363.2 ENSMUST00000114705.9 ENSMUST00000002100.8 |
Tmem25
|
transmembrane protein 25 |
| chr1_+_66739990 | 1.30 |
ENSMUST00000027157.10
ENSMUST00000113995.2 |
Rpe
|
ribulose-5-phosphate-3-epimerase |
| chr9_-_65815958 | 1.30 |
ENSMUST00000119245.8
ENSMUST00000134338.8 ENSMUST00000179395.8 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr10_+_79726687 | 1.30 |
ENSMUST00000217837.2
ENSMUST00000061653.9 |
Cfd
|
complement factor D (adipsin) |
| chr4_+_141095415 | 1.29 |
ENSMUST00000006380.5
|
Fam131c
|
family with sequence similarity 131, member C |
| chr19_+_11514132 | 1.29 |
ENSMUST00000025581.7
|
Ms4a4d
|
membrane-spanning 4-domains, subfamily A, member 4D |
| chr7_-_14226851 | 1.29 |
ENSMUST00000108524.4
ENSMUST00000211740.2 ENSMUST00000209744.2 |
Sult2a7
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 7 |
| chr6_+_149043136 | 1.29 |
ENSMUST00000166416.8
ENSMUST00000111551.2 |
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr19_+_46695889 | 1.29 |
ENSMUST00000003655.9
|
As3mt
|
arsenite methyltransferase |
| chr14_-_68771138 | 1.28 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr6_+_129157576 | 1.28 |
ENSMUST00000032260.6
|
Clec2d
|
C-type lectin domain family 2, member d |
| chr3_-_98660781 | 1.28 |
ENSMUST00000094050.11
ENSMUST00000090743.13 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr15_+_3300249 | 1.27 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr12_-_103623354 | 1.27 |
ENSMUST00000152517.2
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr7_-_30428930 | 1.27 |
ENSMUST00000207296.2
ENSMUST00000006478.10 |
Tmem147
|
transmembrane protein 147 |
| chr9_+_7692087 | 1.27 |
ENSMUST00000018767.8
|
Mmp7
|
matrix metallopeptidase 7 |
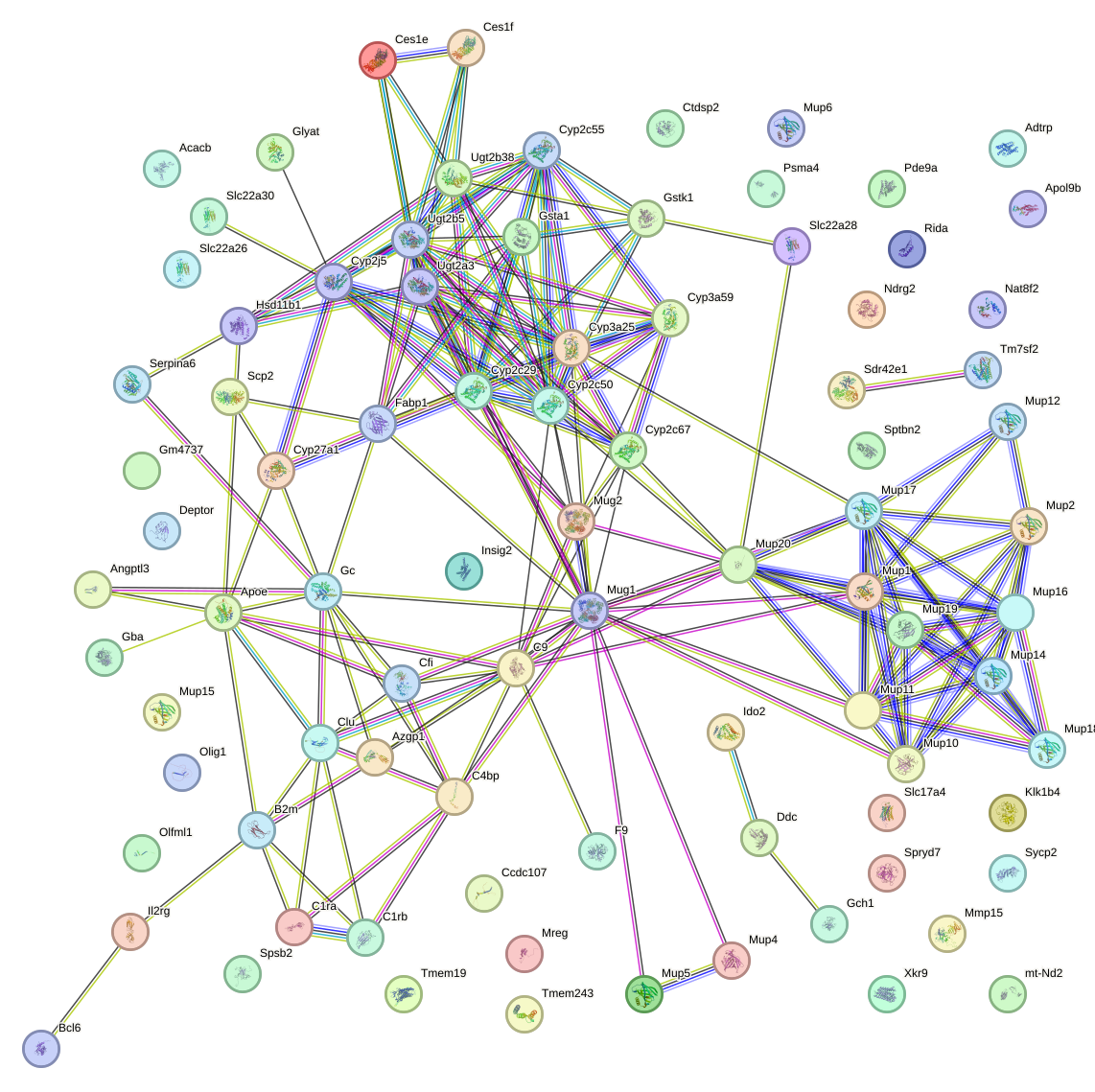
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 2.3 | 27.7 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.9 | 11.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 1.8 | 22.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 1.7 | 5.2 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 1.7 | 18.2 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 1.6 | 23.7 | GO:0015747 | urate transport(GO:0015747) |
| 1.6 | 3.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 1.5 | 4.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.3 | 5.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.2 | 7.1 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.1 | 3.4 | GO:1904735 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 1.0 | 5.8 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.9 | 3.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.9 | 2.8 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.8 | 3.4 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.8 | 2.5 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.8 | 2.4 | GO:1903989 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.7 | 5.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.7 | 2.2 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.7 | 4.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.6 | 1.9 | GO:0019255 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.6 | 11.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.5 | 2.1 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.5 | 1.5 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.5 | 2.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.5 | 6.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.5 | 2.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.5 | 8.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 0.5 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.5 | 1.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 1.8 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.5 | 2.3 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.4 | 1.3 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.4 | 1.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.4 | 1.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.4 | 1.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.4 | 1.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.4 | 1.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.4 | 1.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.4 | 1.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.3 | 2.0 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.3 | 1.0 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 0.3 | 2.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 1.3 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.3 | 0.9 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.3 | 5.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 2.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.3 | 1.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 2.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.3 | 1.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.3 | 2.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 0.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.3 | 2.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.3 | 0.9 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.3 | 0.9 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.3 | 6.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 1.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.3 | 1.1 | GO:0070781 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.3 | 7.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 0.8 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 0.8 | GO:0046491 | L-methylmalonyl-CoA metabolic process(GO:0046491) |
| 0.3 | 0.8 | GO:2000097 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 1.5 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.3 | 0.8 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.3 | 1.0 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 1.0 | GO:0050823 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 0.7 | GO:0097037 | heme export(GO:0097037) |
| 0.2 | 0.7 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.2 | 1.9 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 1.4 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.2 | 1.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 1.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.9 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 2.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 1.4 | GO:0015867 | ATP transport(GO:0015867) |
| 0.2 | 0.7 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.2 | 0.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 0.6 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.2 | 0.4 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 4.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.8 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 | 1.2 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 0.6 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.2 | 1.3 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.2 | 0.9 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 | 3.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.9 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 0.5 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 1.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 0.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 1.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 2.8 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.2 | 0.5 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 1.0 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 0.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 1.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 0.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.2 | 1.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.2 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 2.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 2.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 1.7 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.2 | 4.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 1.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.2 | 1.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 5.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.2 | 4.6 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.2 | 0.9 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 1.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 0.8 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.2 | 1.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.6 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 0.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 2.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 3.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 7.5 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.9 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.1 | 1.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.6 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 2.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.4 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 1.8 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.4 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 1.3 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 2.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.8 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.5 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.1 | 2.0 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 0.4 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 0.1 | 2.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.4 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 1.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 1.4 | GO:1902739 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 0.6 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.4 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.9 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 1.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 1.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.7 | GO:0032827 | peptidoglycan metabolic process(GO:0000270) natural killer cell differentiation involved in immune response(GO:0002325) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.4 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.4 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 1.2 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.4 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.1 | 0.7 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.7 | GO:0090204 | protein localization to nuclear pore(GO:0090204) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.5 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.3 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.9 | GO:0043519 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 | 0.3 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 | 1.4 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.1 | 2.0 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.1 | 0.9 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:1902336 | positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.4 | GO:0015755 | fructose transport(GO:0015755) |
| 0.1 | 1.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 4.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.1 | GO:0060529 | cloacal septation(GO:0060197) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.1 | 0.5 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.7 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.4 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.8 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 2.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 1.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.6 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 2.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 0.8 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 2.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.5 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.3 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 0.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.9 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.1 | 0.6 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.3 | GO:0043380 | defense response to nematode(GO:0002215) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.6 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.5 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.1 | 2.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.0 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.6 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.1 | 1.0 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 2.6 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 2.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 2.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.2 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.3 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 1.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.6 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.5 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 0.5 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.4 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.7 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.2 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.1 | 0.7 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.4 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 2.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.1 | 0.5 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 2.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.2 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of cardiac ventricle development(GO:1904412) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.1 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.5 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.6 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.9 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.2 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 8.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.4 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.6 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.1 | 0.3 | GO:1900738 | psychomotor behavior(GO:0036343) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.9 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.1 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.3 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.0 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.3 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.6 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.1 | 1.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.5 | GO:0098762 | meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.1 | 0.6 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.5 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.1 | 0.3 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 1.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 1.8 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 1.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.4 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.6 | GO:0071554 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.3 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.3 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.3 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 1.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.2 | GO:2000292 | positive regulation of serotonin secretion(GO:0014064) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 1.1 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 2.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0090151 | establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 1.0 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.5 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.8 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:1903294 | positive regulation of glutamate neurotransmitter secretion in response to membrane depolarization(GO:0061646) regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.0 | 0.6 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.0 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 3.2 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.8 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.4 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) primary lung bud formation(GO:0060431) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.1 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 5.1 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.5 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.3 | GO:0060768 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.4 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.8 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.8 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.7 | GO:0071420 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.9 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.3 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.5 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.5 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 2.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.3 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 3.6 | GO:1902652 | secondary alcohol metabolic process(GO:1902652) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 1.4 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.4 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.0 | 0.1 | GO:0010841 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.0 | 1.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 3.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0051446 | positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 1.8 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 1.4 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.8 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.3 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.5 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.2 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.3 | GO:0034341 | response to interferon-gamma(GO:0034341) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 2.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.4 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0060406 | regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0071442 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.5 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.3 | GO:0009583 | detection of light stimulus(GO:0009583) |
| 0.0 | 0.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.6 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 3.2 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.3 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.8 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.0 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.5 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.1 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 1.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.0 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.5 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 1.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.3 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.0 | 0.1 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.0 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 10.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.0 | 5.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.8 | 2.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.8 | 7.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 1.6 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.5 | 2.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.4 | 3.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.4 | 5.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 2.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 1.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 0.9 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.3 | 1.4 | GO:0000802 | transverse filament(GO:0000802) |
| 0.3 | 2.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 1.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 0.7 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.2 | 6.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 2.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.8 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 0.5 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.2 | 0.9 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 0.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 17.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 1.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 1.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.7 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 1.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.7 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 1.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.9 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 2.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 2.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 2.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 8.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 17.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.5 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 2.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.4 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.1 | 3.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 9.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 1.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 6.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 4.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.3 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 2.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 2.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 5.3 | 21.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 4.4 | 22.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 2.9 | 11.5 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 2.6 | 13.2 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 2.5 | 10.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.7 | 5.2 | GO:0046911 | metal chelating activity(GO:0046911) |
| 1.5 | 4.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.4 | 23.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.2 | 4.9 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.0 | 4.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.0 | 3.1 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.9 | 5.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.7 | 2.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.7 | 2.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.6 | 3.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.6 | 3.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.6 | 8.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.5 | 2.1 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.5 | 1.6 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.5 | 2.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 2.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 1.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.4 | 2.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 1.7 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.4 | 2.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 2.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.4 | 1.2 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.4 | 1.6 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.4 | 2.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.4 | 15.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 1.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.4 | 11.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 1.1 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.3 | 3.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.3 | 1.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.3 | 1.2 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.3 | 7.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 6.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.3 | 1.2 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.3 | 0.9 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.3 | 1.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.3 | 1.5 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.3 | 0.8 | GO:0004794 | L-serine ammonia-lyase activity(GO:0003941) L-threonine ammonia-lyase activity(GO:0004794) |
| 0.3 | 2.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.3 | 3.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 1.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.3 | 1.1 | GO:0004077 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.3 | 1.9 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.3 | 2.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 2.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 0.8 | GO:0004493 | methylmalonyl-CoA epimerase activity(GO:0004493) |
| 0.3 | 0.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 2.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 7.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 1.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.2 | 0.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 1.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 1.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 1.1 | GO:0030977 | taurine binding(GO:0030977) |
| 0.2 | 0.7 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.2 | 0.7 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.2 | 2.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 5.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 1.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 4.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 1.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 0.6 | GO:0071820 | N-box binding(GO:0071820) |
| 0.2 | 2.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 0.7 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.2 | 0.9 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 0.5 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 1.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 1.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.2 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 0.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 13.4 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.2 | 0.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 0.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 0.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 5.6 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.2 | 0.6 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.2 | 1.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.0 | GO:0050051 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 21.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 2.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 0.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 2.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 1.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.4 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.7 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 1.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 2.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 2.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.3 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 11.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.5 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.8 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 2.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.7 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.4 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 0.5 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.4 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.3 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 1.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 1.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 2.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 2.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 2.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.5 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 7.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 0.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 1.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 2.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 3.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 1.1 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.4 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 3.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 2.3 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.6 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 1.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 1.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.0 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 2.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 4.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 6.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 5.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 3.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 18.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.8 | 2.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.4 | 5.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 12.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 5.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 8.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 5.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 3.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 6.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 6.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 3.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 1.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 10.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 5.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.7 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 8.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.9 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.1 | 1.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 8.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |