Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxb3
Z-value: 0.53
Transcription factors associated with Hoxb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb3
|
ENSMUSG00000048763.12 | homeobox B3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb3 | mm39_v1_chr11_+_96214078_96214152 | -0.67 | 7.2e-06 | Click! |
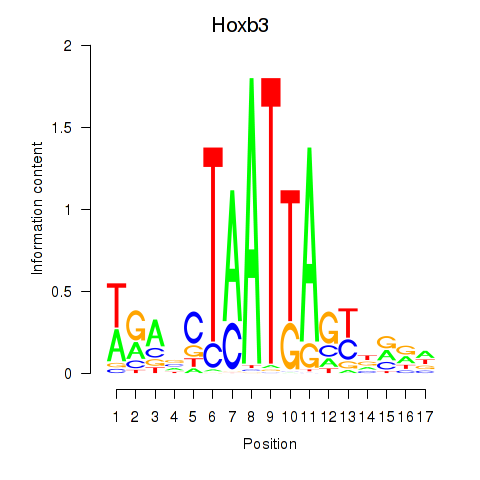
Activity profile of Hoxb3 motif
Sorted Z-values of Hoxb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_60455331 | 4.88 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr5_-_87240405 | 2.24 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr6_-_125357756 | 2.03 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr7_-_14226851 | 1.57 |
ENSMUST00000108524.4
ENSMUST00000211740.2 ENSMUST00000209744.2 |
Sult2a7
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 7 |
| chr1_-_150341911 | 1.53 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr17_+_85335775 | 1.51 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr2_-_154916367 | 1.24 |
ENSMUST00000137242.2
ENSMUST00000054607.16 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chr16_+_22676589 | 1.16 |
ENSMUST00000004574.14
ENSMUST00000178320.2 ENSMUST00000166487.10 |
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr2_-_25351106 | 1.11 |
ENSMUST00000114261.9
|
Paxx
|
non-homologous end joining factor |
| chr7_-_119122681 | 0.99 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr7_+_51528788 | 0.95 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chrX_+_106299484 | 0.80 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr14_-_70666513 | 0.77 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr13_-_21900313 | 0.76 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr17_+_46807637 | 0.75 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr7_-_12829100 | 0.74 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr14_-_70666821 | 0.65 |
ENSMUST00000226229.2
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr18_+_12874390 | 0.64 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr9_-_50472620 | 0.57 |
ENSMUST00000217236.2
|
Tex12
|
testis expressed 12 |
| chr10_+_115854118 | 0.55 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr2_-_132089667 | 0.55 |
ENSMUST00000110163.8
ENSMUST00000180286.2 ENSMUST00000028816.9 |
Tmem230
|
transmembrane protein 230 |
| chr1_+_182392577 | 0.52 |
ENSMUST00000048941.14
|
Capn8
|
calpain 8 |
| chr18_+_36868084 | 0.51 |
ENSMUST00000007046.9
ENSMUST00000237870.2 |
Tmco6
|
transmembrane and coiled-coil domains 6 |
| chrM_+_10167 | 0.50 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr11_-_115310743 | 0.49 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr6_+_68518603 | 0.49 |
ENSMUST00000168090.3
ENSMUST00000103326.3 |
Igkv1-99
|
immunoglobulin kappa variable 1-99 |
| chr2_-_5680801 | 0.49 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr13_-_58506890 | 0.49 |
ENSMUST00000225388.2
|
Kif27
|
kinesin family member 27 |
| chr5_-_143279378 | 0.48 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr17_-_45970238 | 0.46 |
ENSMUST00000120717.8
|
Capn11
|
calpain 11 |
| chr17_+_28491085 | 0.45 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr2_-_17465410 | 0.44 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr9_-_15212849 | 0.44 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr9_-_50472605 | 0.42 |
ENSMUST00000034568.7
|
Tex12
|
testis expressed 12 |
| chr11_+_101556367 | 0.42 |
ENSMUST00000039388.3
|
Arl4d
|
ADP-ribosylation factor-like 4D |
| chrX_+_56257374 | 0.41 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr17_+_88748139 | 0.41 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chr4_-_14621805 | 0.41 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr7_-_24423715 | 0.41 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr5_+_134961246 | 0.41 |
ENSMUST00000111218.8
ENSMUST00000136246.5 ENSMUST00000201847.3 |
Mettl27
|
methyltransferase like 27 |
| chr9_+_21634779 | 0.40 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr7_+_28869770 | 0.40 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr11_-_95966407 | 0.39 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr2_+_151947444 | 0.39 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr18_+_12466876 | 0.39 |
ENSMUST00000092070.13
|
Lama3
|
laminin, alpha 3 |
| chr4_-_14621669 | 0.37 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr18_+_12874368 | 0.37 |
ENSMUST00000235000.2
ENSMUST00000115857.9 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr14_-_52704952 | 0.36 |
ENSMUST00000206520.3
|
Olfr1508
|
olfactory receptor 1508 |
| chr14_+_101891416 | 0.36 |
ENSMUST00000002289.8
|
Uchl3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr10_+_75983285 | 0.36 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr5_-_137015683 | 0.36 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr2_+_69727563 | 0.36 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr17_-_50600620 | 0.35 |
ENSMUST00000010736.9
|
Dazl
|
deleted in azoospermia-like |
| chr6_-_41752111 | 0.35 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr7_+_28869629 | 0.35 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr3_-_105921946 | 0.35 |
ENSMUST00000010280.11
|
Pifo
|
primary cilia formation |
| chr14_-_44987797 | 0.34 |
ENSMUST00000226900.2
|
Gm8267
|
predicted gene 8267 |
| chr18_+_34675366 | 0.34 |
ENSMUST00000012426.3
|
Wnt8a
|
wingless-type MMTV integration site family, member 8A |
| chrX_-_138683102 | 0.34 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr17_+_46608333 | 0.33 |
ENSMUST00000188223.7
ENSMUST00000061722.13 ENSMUST00000166280.8 |
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr11_+_116734104 | 0.33 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr7_-_101749433 | 0.33 |
ENSMUST00000106937.8
|
Art5
|
ADP-ribosyltransferase 5 |
| chr13_-_21726945 | 0.33 |
ENSMUST00000205976.3
ENSMUST00000175637.3 |
Olfr1366
|
olfactory receptor 1366 |
| chr9_-_105272435 | 0.33 |
ENSMUST00000140851.3
|
Nek11
|
NIMA (never in mitosis gene a)-related expressed kinase 11 |
| chr2_+_127696548 | 0.33 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr12_+_84332006 | 0.32 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chr5_+_134961205 | 0.32 |
ENSMUST00000047196.14
ENSMUST00000111221.9 ENSMUST00000111219.8 ENSMUST00000068617.12 |
Mettl27
|
methyltransferase like 27 |
| chr4_+_3938881 | 0.32 |
ENSMUST00000108386.8
ENSMUST00000121110.8 ENSMUST00000149544.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr11_-_94673526 | 0.32 |
ENSMUST00000100554.8
|
Tmem92
|
transmembrane protein 92 |
| chr19_-_12302465 | 0.32 |
ENSMUST00000207241.3
|
Olfr1437
|
olfactory receptor 1437 |
| chr7_+_4925781 | 0.31 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr6_-_68968278 | 0.31 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr7_-_24371457 | 0.31 |
ENSMUST00000078001.7
|
Tex101
|
testis expressed gene 101 |
| chr11_-_95966477 | 0.31 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr14_-_36820304 | 0.30 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr5_-_143963413 | 0.30 |
ENSMUST00000031622.13
|
Ocm
|
oncomodulin |
| chr16_-_58695131 | 0.30 |
ENSMUST00000217377.2
|
Olfr177
|
olfactory receptor 177 |
| chr10_-_128885867 | 0.30 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr17_-_45785752 | 0.30 |
ENSMUST00000233553.2
ENSMUST00000233769.2 ENSMUST00000024731.9 |
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr10_+_128173603 | 0.29 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr14_+_53521353 | 0.28 |
ENSMUST00000103625.3
|
Trav3n-3
|
T cell receptor alpha variable 3N-3 |
| chr8_+_114362419 | 0.28 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr4_+_3938903 | 0.28 |
ENSMUST00000121210.8
ENSMUST00000121651.8 ENSMUST00000041122.11 ENSMUST00000120732.8 ENSMUST00000119307.8 ENSMUST00000123769.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr13_-_53627110 | 0.28 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr2_-_69542805 | 0.28 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr2_-_86208737 | 0.28 |
ENSMUST00000217435.2
|
Olfr1057
|
olfactory receptor 1057 |
| chr5_-_74692327 | 0.27 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr19_+_60800012 | 0.27 |
ENSMUST00000128357.8
ENSMUST00000119633.8 ENSMUST00000025957.9 |
Dennd10
|
DENN domain containing 10 |
| chr4_-_144513121 | 0.27 |
ENSMUST00000105746.3
|
Aadacl4fm5
|
AADACL4 family member 5 |
| chr18_+_52748978 | 0.26 |
ENSMUST00000072666.4
ENSMUST00000209270.2 |
Zfp474
|
zinc finger protein 474 |
| chr9_-_99302205 | 0.26 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr7_-_103420801 | 0.25 |
ENSMUST00000106878.3
|
Olfr69
|
olfactory receptor 69 |
| chr12_+_101370932 | 0.25 |
ENSMUST00000055156.5
|
Catsperb
|
cation channel sperm associated auxiliary subunit beta |
| chr13_-_112698563 | 0.25 |
ENSMUST00000224510.2
|
Il31ra
|
interleukin 31 receptor A |
| chr11_+_94632608 | 0.25 |
ENSMUST00000021240.7
ENSMUST00000188741.2 |
Cdc34b
|
cell division cycle 34B |
| chr10_+_29074950 | 0.25 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr2_-_157188568 | 0.25 |
ENSMUST00000029172.2
|
Ghrh
|
growth hormone releasing hormone |
| chr15_-_8739893 | 0.25 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr16_+_51851917 | 0.24 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr6_-_130106861 | 0.24 |
ENSMUST00000014476.6
|
Klra8
|
killer cell lectin-like receptor, subfamily A, member 8 |
| chr4_-_114991174 | 0.24 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr1_-_92412835 | 0.24 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chr12_-_81579614 | 0.24 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr12_-_99529767 | 0.23 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr10_+_129539079 | 0.23 |
ENSMUST00000213331.2
|
Olfr804
|
olfactory receptor 804 |
| chr8_-_105261262 | 0.23 |
ENSMUST00000162466.8
ENSMUST00000034349.10 |
Nae1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr5_-_137213788 | 0.23 |
ENSMUST00000239135.2
|
Gm31160
|
predicted gene, 31160 |
| chr16_+_51851948 | 0.23 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr3_+_31956656 | 0.23 |
ENSMUST00000119310.8
|
Kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr1_+_54289833 | 0.23 |
ENSMUST00000027128.11
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr2_+_3425159 | 0.23 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr11_-_73800125 | 0.23 |
ENSMUST00000215690.2
|
Olfr395
|
olfactory receptor 395 |
| chr10_-_128904846 | 0.22 |
ENSMUST00000204515.3
|
Olfr766-ps1
|
olfactory receptor 766, pseudogene 1 |
| chr9_+_18320390 | 0.22 |
ENSMUST00000098973.3
|
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr2_+_36575800 | 0.22 |
ENSMUST00000213258.2
|
Olfr346
|
olfactory receptor 346 |
| chr2_-_111324108 | 0.22 |
ENSMUST00000208881.2
ENSMUST00000208695.2 ENSMUST00000217611.2 |
Olfr1290
|
olfactory receptor 1290 |
| chr1_+_173876771 | 0.22 |
ENSMUST00000213381.2
ENSMUST00000213211.2 |
Olfr432
|
olfactory receptor 432 |
| chr7_-_114235506 | 0.22 |
ENSMUST00000205714.2
ENSMUST00000206853.2 ENSMUST00000205933.2 ENSMUST00000206156.2 ENSMUST00000032907.9 ENSMUST00000032906.11 |
Calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chrY_+_5656986 | 0.22 |
ENSMUST00000190391.2
|
Gm21854
|
predicted gene, 21854 |
| chr9_+_40092216 | 0.22 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr10_-_75946790 | 0.22 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr18_+_61408073 | 0.21 |
ENSMUST00000135688.2
|
Pde6a
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr7_+_6441687 | 0.21 |
ENSMUST00000218906.2
|
Olfr1344
|
olfactory receptor 1344 |
| chr15_+_10952418 | 0.21 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr10_+_127919142 | 0.21 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr7_+_114342929 | 0.21 |
ENSMUST00000161800.2
|
Insc
|
INSC spindle orientation adaptor protein |
| chr2_-_89951611 | 0.21 |
ENSMUST00000216493.2
ENSMUST00000214404.2 |
Olfr1269
|
olfactory receptor 1269 |
| chr8_+_114362181 | 0.21 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr12_+_74044435 | 0.20 |
ENSMUST00000221220.2
|
Syt16
|
synaptotagmin XVI |
| chr7_+_103652466 | 0.20 |
ENSMUST00000209757.3
|
Olfr638
|
olfactory receptor 638 |
| chrY_+_5942460 | 0.20 |
ENSMUST00000185926.2
|
Gm21778
|
predicted gene, 21778 |
| chr6_-_112364974 | 0.20 |
ENSMUST00000238755.2
ENSMUST00000060847.6 |
Ssu2
|
ssu-2 homolog (C. elegans) |
| chr4_-_14621497 | 0.20 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr16_-_30086317 | 0.20 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr13_-_57043550 | 0.20 |
ENSMUST00000022023.13
ENSMUST00000174457.9 ENSMUST00000109871.8 |
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr1_-_92107971 | 0.19 |
ENSMUST00000186002.3
ENSMUST00000097644.9 |
Hdac4
|
histone deacetylase 4 |
| chr7_-_41098120 | 0.19 |
ENSMUST00000233793.2
ENSMUST00000233555.2 ENSMUST00000165029.3 |
Vmn2r57
|
vomeronasal 2, receptor 57 |
| chr17_-_37591309 | 0.19 |
ENSMUST00000077585.3
|
Olfr99
|
olfactory receptor 99 |
| chr19_-_5610628 | 0.19 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr2_+_118877610 | 0.19 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr14_-_50425655 | 0.19 |
ENSMUST00000205837.3
|
Olfr730
|
olfactory receptor 730 |
| chr6_-_89825033 | 0.19 |
ENSMUST00000226436.2
|
Vmn1r42
|
vomeronasal 1 receptor 42 |
| chr4_-_155729865 | 0.19 |
ENSMUST00000115821.3
|
Gm10563
|
predicted gene 10563 |
| chr1_-_92423800 | 0.18 |
ENSMUST00000204766.2
ENSMUST00000204009.3 |
Olfr1415
|
olfactory receptor 1415 |
| chr6_-_89853395 | 0.18 |
ENSMUST00000227279.2
ENSMUST00000228709.2 ENSMUST00000226983.2 |
Vmn1r42
Vmn1r43
|
vomeronasal 1 receptor 42 vomeronasal 1 receptor 43 |
| chr18_+_52958382 | 0.18 |
ENSMUST00000238707.2
|
Gm50457
|
predicted gene, 50457 |
| chr1_-_9369045 | 0.18 |
ENSMUST00000191683.6
|
Sntg1
|
syntrophin, gamma 1 |
| chr7_-_139734637 | 0.18 |
ENSMUST00000059241.8
|
Sprn
|
shadow of prion protein |
| chr4_-_3938352 | 0.18 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr1_+_135768409 | 0.17 |
ENSMUST00000189826.7
|
Tnnt2
|
troponin T2, cardiac |
| chr1_+_172383499 | 0.17 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr13_-_23929490 | 0.17 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr14_-_34322093 | 0.17 |
ENSMUST00000022331.3
|
Opn4
|
opsin 4 (melanopsin) |
| chr11_+_95275458 | 0.17 |
ENSMUST00000021243.16
ENSMUST00000146556.2 |
Slc35b1
|
solute carrier family 35, member B1 |
| chr8_+_22329942 | 0.17 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chr9_-_75504926 | 0.17 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chr10_+_97442727 | 0.17 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chr11_-_34158085 | 0.17 |
ENSMUST00000060271.3
|
Foxi1
|
forkhead box I1 |
| chr7_-_12787611 | 0.16 |
ENSMUST00000182490.2
|
Mzf1
|
myeloid zinc finger 1 |
| chrM_+_8603 | 0.16 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr7_-_86016045 | 0.16 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr13_+_38388904 | 0.16 |
ENSMUST00000091641.13
ENSMUST00000178564.2 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr10_+_102210290 | 0.16 |
ENSMUST00000120748.2
|
Mgat4c
|
MGAT4 family, member C |
| chr14_+_8555284 | 0.16 |
ENSMUST00000144914.3
|
Gm281
|
predicted gene 281 |
| chr6_+_90278195 | 0.15 |
ENSMUST00000054799.9
|
BC048671
|
cDNA sequence BC048671 |
| chr9_+_35819708 | 0.15 |
ENSMUST00000176049.2
ENSMUST00000176153.2 |
Pate13
|
prostate and testis expressed 13 |
| chr2_-_86257093 | 0.15 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr1_-_9368721 | 0.15 |
ENSMUST00000132064.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr6_-_23650205 | 0.15 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr14_+_62901114 | 0.15 |
ENSMUST00000171692.2
|
Serpine3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr2_+_68490186 | 0.15 |
ENSMUST00000055930.6
|
4932414N04Rik
|
RIKEN cDNA 4932414N04 gene |
| chr9_+_119170486 | 0.15 |
ENSMUST00000175743.8
ENSMUST00000176397.8 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr11_-_102076028 | 0.15 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chr3_+_65573644 | 0.15 |
ENSMUST00000099075.4
ENSMUST00000177434.3 |
Lekr1
|
leucine, glutamate and lysine rich 1 |
| chr11_+_49327451 | 0.14 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr11_-_59484115 | 0.14 |
ENSMUST00000215626.2
|
Olfr223
|
olfactory receptor 223 |
| chr12_-_25146078 | 0.14 |
ENSMUST00000222667.2
ENSMUST00000020974.7 |
Id2
|
inhibitor of DNA binding 2 |
| chr2_+_87854404 | 0.14 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr16_-_22676264 | 0.14 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr15_-_3333003 | 0.14 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chrX_-_9123138 | 0.14 |
ENSMUST00000115553.3
|
Gm14862
|
predicted gene 14862 |
| chr9_-_15212745 | 0.14 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr2_+_69727599 | 0.14 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_-_164480710 | 0.13 |
ENSMUST00000109336.2
|
Wfdc16
|
WAP four-disulfide core domain 16 |
| chrX_-_49108236 | 0.13 |
ENSMUST00000213556.3
ENSMUST00000213463.2 |
Olfr1323
|
olfactory receptor 1323 |
| chr17_+_38143840 | 0.13 |
ENSMUST00000213857.2
|
Olfr125
|
olfactory receptor 125 |
| chr2_-_160950936 | 0.13 |
ENSMUST00000039782.14
ENSMUST00000134178.8 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr10_-_128918779 | 0.13 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr19_+_12364643 | 0.13 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chr14_+_50360643 | 0.13 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr7_-_107696793 | 0.13 |
ENSMUST00000217304.2
|
Olfr482
|
olfactory receptor 482 |
| chr6_-_68887922 | 0.13 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr6_-_68784692 | 0.13 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr6_-_69020489 | 0.13 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr11_+_73489420 | 0.12 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr14_+_25979825 | 0.12 |
ENSMUST00000173580.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr4_+_62443606 | 0.12 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr12_-_111150925 | 0.12 |
ENSMUST00000121608.2
|
4930595D18Rik
|
RIKEN cDNA 4930595D18 gene |
| chr6_+_40419797 | 0.12 |
ENSMUST00000038907.9
ENSMUST00000141490.2 |
Wee2
|
WEE1 homolog 2 (S. pombe) |
| chr11_-_45845992 | 0.12 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
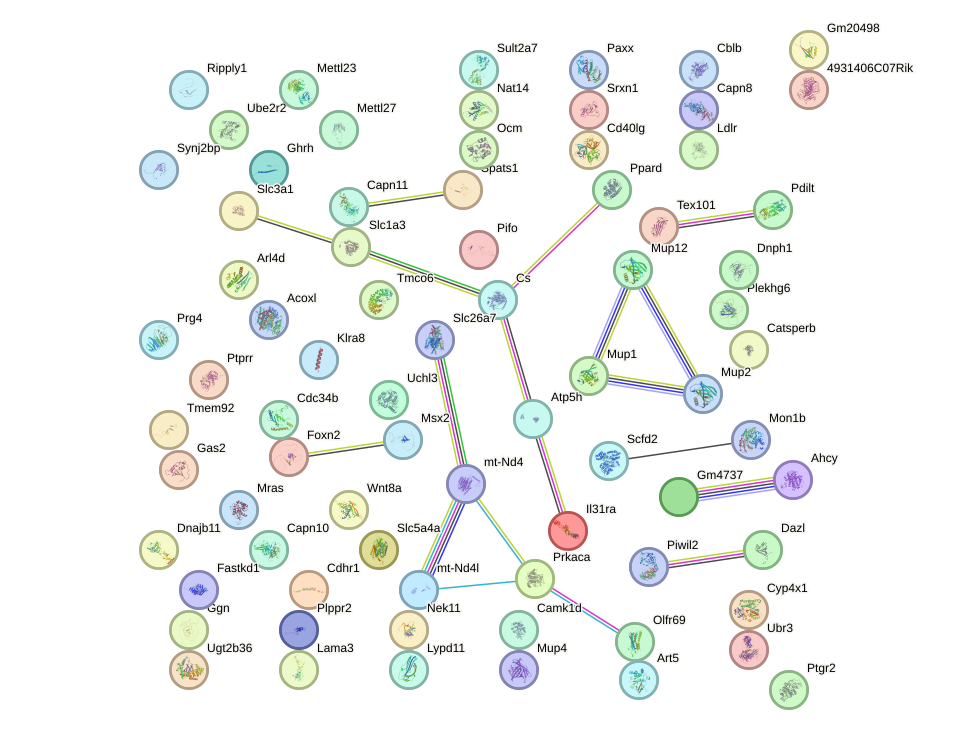
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.9 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.4 | 1.4 | GO:0000239 | pachytene(GO:0000239) |
| 0.3 | 1.2 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.2 | 1.5 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 1.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 0.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.4 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.4 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.3 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.3 | GO:1902336 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.1 | 0.7 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 1.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.2 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.6 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.3 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.1 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.6 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 1.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.0 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.2 | GO:0014870 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 1.0 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 0.4 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.2 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.4 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 1.0 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 2.5 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.8 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.2 | 1.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.4 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 1.0 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.3 | 1.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 1.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 1.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.3 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 2.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 1.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.0 | 0.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.8 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.7 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |