Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxb6
Z-value: 0.32
Transcription factors associated with Hoxb6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb6
|
ENSMUSG00000000690.6 | homeobox B6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb6 | mm39_v1_chr11_+_96183294_96183334 | 0.25 | 1.5e-01 | Click! |
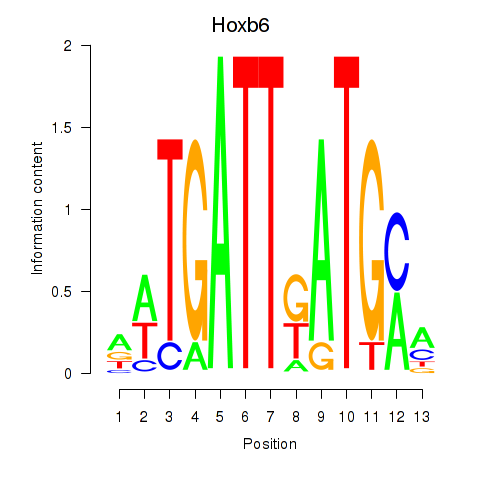
Activity profile of Hoxb6 motif
Sorted Z-values of Hoxb6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_164611812 | 3.97 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr15_+_34306812 | 1.40 |
ENSMUST00000226766.2
ENSMUST00000163455.9 ENSMUST00000022947.7 ENSMUST00000228570.2 ENSMUST00000227759.2 |
Matn2
|
matrilin 2 |
| chr6_+_86348286 | 1.28 |
ENSMUST00000089558.7
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr10_-_117628565 | 1.23 |
ENSMUST00000167943.8
ENSMUST00000064848.7 |
Nup107
|
nucleoporin 107 |
| chr13_-_22017677 | 1.09 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr3_-_100876960 | 0.88 |
ENSMUST00000076941.12
|
Ttf2
|
transcription termination factor, RNA polymerase II |
| chr6_+_142244145 | 0.79 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr11_-_96868483 | 0.79 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor |
| chr2_+_85809620 | 0.77 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr16_-_19079594 | 0.71 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr7_-_115459082 | 0.68 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr11_-_99134885 | 0.64 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr16_-_18904240 | 0.63 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr2_+_29236815 | 0.54 |
ENSMUST00000028139.11
ENSMUST00000113830.11 |
Med27
|
mediator complex subunit 27 |
| chrX_-_100266032 | 0.50 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr9_-_56151334 | 0.47 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr3_+_94320548 | 0.47 |
ENSMUST00000166032.8
ENSMUST00000200486.5 ENSMUST00000196386.5 ENSMUST00000045245.10 ENSMUST00000197901.5 ENSMUST00000198041.2 |
Tdrkh
Gm42463
|
tudor and KH domain containing protein predicted gene 42463 |
| chrX_-_8118541 | 0.46 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr16_+_44167484 | 0.45 |
ENSMUST00000050897.7
|
Spice1
|
spindle and centriole associated protein 1 |
| chr8_-_106198112 | 0.43 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_-_138103021 | 0.42 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr11_-_74615496 | 0.40 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr13_+_65298054 | 0.40 |
ENSMUST00000214214.2
|
Olfr466
|
olfactory receptor 466 |
| chr14_-_57353340 | 0.38 |
ENSMUST00000159455.2
|
Gm4491
|
predicted gene 4491 |
| chr1_-_36312482 | 0.37 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr9_-_35481689 | 0.31 |
ENSMUST00000115110.5
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr7_-_28661648 | 0.25 |
ENSMUST00000127210.8
|
Actn4
|
actinin alpha 4 |
| chr3_+_138058139 | 0.25 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr8_+_31601837 | 0.25 |
ENSMUST00000046941.8
ENSMUST00000217278.2 |
Rnf122
|
ring finger protein 122 |
| chr1_-_138102972 | 0.24 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr1_-_179572765 | 0.24 |
ENSMUST00000211943.3
ENSMUST00000131716.4 ENSMUST00000221136.2 |
Kif28
|
kinesin family member 28 |
| chr5_-_123804745 | 0.24 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr1_+_106866678 | 0.23 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr3_+_64884839 | 0.21 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr9_+_74959259 | 0.21 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr11_+_49410475 | 0.20 |
ENSMUST00000204706.3
|
Olfr1383
|
olfactory receptor 1383 |
| chr2_-_88157559 | 0.19 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr3_-_79053182 | 0.16 |
ENSMUST00000118340.7
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr11_-_72106418 | 0.15 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr2_-_125701059 | 0.14 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chrX_+_162945162 | 0.14 |
ENSMUST00000131543.2
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr4_-_52919172 | 0.14 |
ENSMUST00000107667.3
ENSMUST00000213989.2 |
Olfr272
|
olfactory receptor 272 |
| chrX_+_106132055 | 0.14 |
ENSMUST00000150494.2
|
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr18_+_37864045 | 0.12 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr17_+_18518361 | 0.11 |
ENSMUST00000231938.2
ENSMUST00000079206.8 ENSMUST00000231879.3 |
Vmn2r93
|
vomeronasal 2, receptor 93 |
| chr3_+_66892979 | 0.10 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr4_+_151081538 | 0.10 |
ENSMUST00000030803.2
|
Uts2
|
urotensin 2 |
| chrX_+_56257374 | 0.08 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr7_-_28661751 | 0.07 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr16_+_58967409 | 0.07 |
ENSMUST00000216957.3
|
Olfr195
|
olfactory receptor 195 |
| chr2_-_86589298 | 0.07 |
ENSMUST00000215991.2
ENSMUST00000217043.3 |
Olfr1090
|
olfactory receptor 1090 |
| chr11_-_65636651 | 0.07 |
ENSMUST00000138093.2
|
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr6_-_69245427 | 0.06 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr6_-_3494587 | 0.06 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr5_-_31684036 | 0.05 |
ENSMUST00000202421.2
ENSMUST00000201769.4 ENSMUST00000065388.11 |
Supt7l
|
SPT7-like, STAGA complex gamma subunit |
| chr10_+_78816884 | 0.05 |
ENSMUST00000058991.5
ENSMUST00000203973.2 |
Olfr1352
|
olfactory receptor 1352 |
| chr1_+_161222980 | 0.05 |
ENSMUST00000028024.5
|
Tnfsf4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr7_-_38227617 | 0.04 |
ENSMUST00000079759.6
|
Gm5591
|
predicted gene 5591 |
| chr4_+_15881256 | 0.04 |
ENSMUST00000029876.2
|
Calb1
|
calbindin 1 |
| chr5_-_52723607 | 0.03 |
ENSMUST00000199942.5
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr6_+_142359099 | 0.03 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr7_+_108019628 | 0.02 |
ENSMUST00000213521.3
|
Olfr497
|
olfactory receptor 497 |
| chr19_+_4189786 | 0.02 |
ENSMUST00000096338.5
|
Gpr152
|
G protein-coupled receptor 152 |
| chr10_-_38998272 | 0.01 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr4_+_33062999 | 0.01 |
ENSMUST00000108162.8
ENSMUST00000024035.9 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr19_+_46385448 | 0.00 |
ENSMUST00000118440.3
|
Sufu
|
SUFU negative regulator of hedgehog signaling |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.5 | 1.4 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.2 | 1.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 0.5 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 | 0.7 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 0.8 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.9 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.1 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:0043382 | defense response to nematode(GO:0002215) positive regulation of memory T cell differentiation(GO:0043382) negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.5 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 4.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |