Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxc13_Hoxd13
Z-value: 0.61
Transcription factors associated with Hoxc13_Hoxd13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
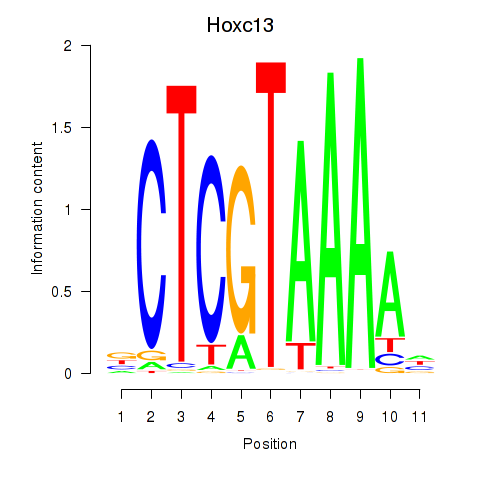
Hoxc13
|
ENSMUSG00000001655.7 | homeobox C13 |
|
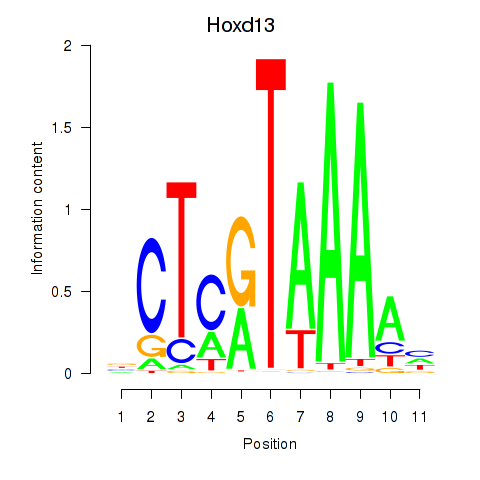
Hoxd13
|
ENSMUSG00000001819.5 | homeobox D13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd13 | mm39_v1_chr2_+_74498551_74498654 | -0.14 | 4.3e-01 | Click! |
Activity profile of Hoxc13_Hoxd13 motif
Sorted Z-values of Hoxc13_Hoxd13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_38342815 | 2.95 |
ENSMUST00000057185.13
|
Pcdh1
|
protocadherin 1 |
| chr4_-_141553306 | 2.33 |
ENSMUST00000102481.4
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr17_-_31363245 | 2.31 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr5_-_66238313 | 2.04 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr10_+_87695352 | 1.96 |
ENSMUST00000121952.8
ENSMUST00000126490.8 |
Igf1
|
insulin-like growth factor 1 |
| chr8_+_57004125 | 1.46 |
ENSMUST00000110322.9
ENSMUST00000040218.13 ENSMUST00000210863.2 |
Fbxo8
|
F-box protein 8 |
| chr9_-_51240201 | 1.29 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr8_-_70591799 | 1.27 |
ENSMUST00000075724.9
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr10_+_87695886 | 1.26 |
ENSMUST00000062862.13
|
Igf1
|
insulin-like growth factor 1 |
| chr13_-_63712140 | 1.26 |
ENSMUST00000195756.6
|
Ptch1
|
patched 1 |
| chr7_+_28240262 | 1.19 |
ENSMUST00000119180.4
|
Sycn
|
syncollin |
| chr5_+_114284585 | 1.19 |
ENSMUST00000102582.8
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr10_+_116111441 | 1.19 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr6_+_108190163 | 1.13 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr3_+_79793237 | 1.08 |
ENSMUST00000029567.9
|
Gask1b
|
golgi associated kinase 1B |
| chr14_-_66361931 | 1.08 |
ENSMUST00000070515.2
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr17_+_35658131 | 1.07 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr16_+_34815177 | 1.07 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase |
| chr7_+_67297152 | 1.07 |
ENSMUST00000032774.16
ENSMUST00000107471.8 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr2_-_127634387 | 1.00 |
ENSMUST00000135091.2
|
Mtln
|
mitoregulin |
| chr8_-_70591746 | 0.98 |
ENSMUST00000212320.2
|
Rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr7_-_73187369 | 0.93 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr5_-_122639840 | 0.92 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr6_-_24528012 | 0.86 |
ENSMUST00000023851.9
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr6_+_108190050 | 0.83 |
ENSMUST00000032192.9
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr1_+_181952302 | 0.83 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr10_+_116137277 | 0.76 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr5_+_128677863 | 0.74 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10 |
| chr11_-_53321242 | 0.71 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr16_-_57051829 | 0.71 |
ENSMUST00000023431.8
|
Tbc1d23
|
TBC1 domain family, member 23 |
| chr17_-_13981703 | 0.71 |
ENSMUST00000127032.8
|
Tcte2
|
t-complex-associated testis expressed 2 |
| chr15_-_101336669 | 0.70 |
ENSMUST00000081945.5
|
Krt87
|
keratin 87 |
| chr11_-_5492175 | 0.69 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr5_+_90708962 | 0.62 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr3_+_81904229 | 0.62 |
ENSMUST00000029641.10
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr10_-_120815232 | 0.62 |
ENSMUST00000119944.8
ENSMUST00000119093.2 |
Lemd3
|
LEM domain containing 3 |
| chr11_-_99907030 | 0.61 |
ENSMUST00000018399.3
|
Krt33a
|
keratin 33A |
| chr11_+_96085118 | 0.60 |
ENSMUST00000062709.4
|
Hoxb13
|
homeobox B13 |
| chr16_-_57051727 | 0.59 |
ENSMUST00000226586.2
|
Tbc1d23
|
TBC1 domain family, member 23 |
| chr19_+_18609343 | 0.59 |
ENSMUST00000159572.9
|
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr11_+_96177449 | 0.54 |
ENSMUST00000049352.8
|
Hoxb7
|
homeobox B7 |
| chr11_-_99920694 | 0.54 |
ENSMUST00000073890.4
|
Krt33b
|
keratin 33B |
| chr13_-_17869314 | 0.54 |
ENSMUST00000221598.2
ENSMUST00000068545.6 ENSMUST00000220514.2 |
Sugct
|
succinyl-CoA glutarate-CoA transferase |
| chr18_+_36431732 | 0.53 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chr6_-_52195663 | 0.50 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr4_+_54945038 | 0.47 |
ENSMUST00000133895.8
|
Zfp462
|
zinc finger protein 462 |
| chr10_+_28544556 | 0.46 |
ENSMUST00000161345.2
|
Themis
|
thymocyte selection associated |
| chr16_+_41353360 | 0.43 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr1_+_60948149 | 0.41 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr13_-_51723473 | 0.40 |
ENSMUST00000239056.2
ENSMUST00000223543.3 |
Shc3
|
src homology 2 domain-containing transforming protein C3 |
| chr1_+_107289659 | 0.40 |
ENSMUST00000027566.3
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr12_+_11506053 | 0.40 |
ENSMUST00000124065.2
|
Rad51ap2
|
RAD51 associated protein 2 |
| chr8_+_105991280 | 0.39 |
ENSMUST00000036221.12
|
Fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr5_+_106024398 | 0.38 |
ENSMUST00000150440.8
ENSMUST00000031227.11 |
Zfp326
|
zinc finger protein 326 |
| chr2_+_74528071 | 0.38 |
ENSMUST00000059272.10
|
Hoxd9
|
homeobox D9 |
| chr1_-_138775317 | 0.37 |
ENSMUST00000093486.10
ENSMUST00000046870.13 |
Lhx9
|
LIM homeobox protein 9 |
| chr2_-_111815654 | 0.36 |
ENSMUST00000214537.2
|
Olfr1309
|
olfactory receptor 1309 |
| chr6_+_48425034 | 0.35 |
ENSMUST00000212740.2
ENSMUST00000169350.9 ENSMUST00000043676.12 |
Sspo
|
SCO-spondin |
| chr11_-_99979052 | 0.34 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr12_+_75355082 | 0.34 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr8_-_25528972 | 0.32 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr17_+_8502682 | 0.32 |
ENSMUST00000124023.8
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr15_-_66158445 | 0.32 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3 |
| chr10_-_128532126 | 0.29 |
ENSMUST00000000727.4
|
Rab5b
|
RAB5B, member RAS oncogene family |
| chr5_+_135093056 | 0.29 |
ENSMUST00000071263.7
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr2_-_34261121 | 0.29 |
ENSMUST00000127353.3
ENSMUST00000141653.3 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr5_+_17779273 | 0.28 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr13_-_63712349 | 0.26 |
ENSMUST00000192155.6
|
Ptch1
|
patched 1 |
| chr10_+_28544356 | 0.26 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr5_+_150119860 | 0.25 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr6_+_24528143 | 0.25 |
ENSMUST00000031696.10
|
Asb15
|
ankyrin repeat and SOCS box-containing 15 |
| chr8_+_113296675 | 0.24 |
ENSMUST00000034225.7
ENSMUST00000118171.8 |
Cntnap4
|
contactin associated protein-like 4 |
| chr16_-_89305201 | 0.24 |
ENSMUST00000056118.4
|
Krtap7-1
|
keratin associated protein 7-1 |
| chr11_-_99996452 | 0.23 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr17_+_8502594 | 0.23 |
ENSMUST00000155364.8
ENSMUST00000046754.15 |
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr17_-_52133594 | 0.23 |
ENSMUST00000129667.8
ENSMUST00000169480.8 ENSMUST00000148559.2 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr11_-_99412084 | 0.23 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr1_-_39844467 | 0.22 |
ENSMUST00000171319.4
|
Gm3646
|
predicted gene 3646 |
| chr9_-_110775143 | 0.22 |
ENSMUST00000199782.2
ENSMUST00000035075.13 |
Tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr16_+_23044763 | 0.22 |
ENSMUST00000178797.8
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr15_-_101482320 | 0.21 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr6_-_52194414 | 0.21 |
ENSMUST00000140316.2
|
Hoxa7
|
homeobox A7 |
| chr1_+_53352780 | 0.21 |
ENSMUST00000027265.10
ENSMUST00000114484.8 |
Osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr3_-_88366159 | 0.20 |
ENSMUST00000147200.8
ENSMUST00000169222.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr7_-_28329924 | 0.20 |
ENSMUST00000159095.2
ENSMUST00000159418.8 ENSMUST00000159560.3 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr7_-_30312246 | 0.20 |
ENSMUST00000006476.6
|
Upk1a
|
uroplakin 1A |
| chr4_+_21879660 | 0.19 |
ENSMUST00000029909.3
|
Coq3
|
coenzyme Q3 methyltransferase |
| chr5_+_48140480 | 0.19 |
ENSMUST00000173107.8
ENSMUST00000174313.8 ENSMUST00000174421.8 ENSMUST00000170109.9 |
Slit2
|
slit guidance ligand 2 |
| chr2_-_45001141 | 0.18 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr11_-_99412162 | 0.18 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr8_-_37200051 | 0.18 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr15_+_23036535 | 0.17 |
ENSMUST00000164787.8
|
Cdh18
|
cadherin 18 |
| chr14_+_65903840 | 0.17 |
ENSMUST00000022610.15
|
Scara5
|
scavenger receptor class A, member 5 |
| chr10_-_33920300 | 0.16 |
ENSMUST00000048052.7
|
Calhm4
|
calcium homeostasis modulator family member 4 |
| chr14_+_58308004 | 0.16 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr7_-_28330322 | 0.16 |
ENSMUST00000040112.5
ENSMUST00000239470.2 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr2_-_17465410 | 0.15 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr5_-_106606032 | 0.15 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chr11_+_105885461 | 0.15 |
ENSMUST00000190995.2
|
Ace3
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
| chr11_-_88608958 | 0.14 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr15_-_101441254 | 0.14 |
ENSMUST00000023720.8
|
Krt84
|
keratin 84 |
| chr7_-_30750828 | 0.13 |
ENSMUST00000206341.2
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr7_-_102408573 | 0.13 |
ENSMUST00000210453.3
ENSMUST00000232246.3 ENSMUST00000239110.2 ENSMUST00000060187.15 ENSMUST00000168007.3 |
Olfr560
Olfr78
|
olfactory receptor 560 olfactory receptor 78 |
| chr12_-_34578842 | 0.13 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr18_-_46413886 | 0.12 |
ENSMUST00000236999.2
|
Pggt1b
|
protein geranylgeranyltransferase type I, beta subunit |
| chr11_-_99941369 | 0.12 |
ENSMUST00000007318.2
|
Krt31
|
keratin 31 |
| chr3_-_49711706 | 0.12 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr4_+_134042423 | 0.12 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr8_-_15096046 | 0.12 |
ENSMUST00000050493.4
ENSMUST00000123331.2 |
BB014433
|
expressed sequence BB014433 |
| chr1_+_60948307 | 0.12 |
ENSMUST00000097720.4
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr3_+_126391046 | 0.12 |
ENSMUST00000106401.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr9_-_96601574 | 0.11 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr3_-_75359125 | 0.11 |
ENSMUST00000204341.3
|
Wdr49
|
WD repeat domain 49 |
| chr11_-_23845207 | 0.11 |
ENSMUST00000102863.3
ENSMUST00000020513.10 |
Papolg
|
poly(A) polymerase gamma |
| chr16_+_39804711 | 0.11 |
ENSMUST00000187695.7
|
Lsamp
|
limbic system-associated membrane protein |
| chr9_-_50571080 | 0.10 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr18_+_69478893 | 0.10 |
ENSMUST00000202354.4
|
Tcf4
|
transcription factor 4 |
| chr14_-_118289557 | 0.10 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr10_-_107321938 | 0.09 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chrX_+_111150171 | 0.09 |
ENSMUST00000164272.3
ENSMUST00000132037.2 |
4933403O08Rik
|
RIKEN cDNA 4933403O08 gene |
| chr11_-_99987051 | 0.09 |
ENSMUST00000103127.4
|
Krt35
|
keratin 35 |
| chr5_+_92957231 | 0.09 |
ENSMUST00000113054.9
|
Shroom3
|
shroom family member 3 |
| chr2_+_20727274 | 0.09 |
ENSMUST00000114607.8
|
Etl4
|
enhancer trap locus 4 |
| chr7_-_30750856 | 0.09 |
ENSMUST00000073892.6
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr11_-_102298748 | 0.09 |
ENSMUST00000124755.2
|
Slc25a39
|
solute carrier family 25, member 39 |
| chr17_-_57307018 | 0.09 |
ENSMUST00000002740.3
|
Pspn
|
persephin |
| chr9_+_110948492 | 0.08 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr7_-_133281790 | 0.08 |
ENSMUST00000033278.8
ENSMUST00000122136.2 |
Mmp21
|
matrix metallopeptidase 21 |
| chr5_-_62923463 | 0.08 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chrX_-_135769285 | 0.07 |
ENSMUST00000058814.7
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr11_-_102298281 | 0.07 |
ENSMUST00000107098.8
ENSMUST00000018821.9 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr8_-_43981143 | 0.07 |
ENSMUST00000080135.5
|
Adam26b
|
a disintegrin and metallopeptidase domain 26B |
| chr3_+_102642516 | 0.06 |
ENSMUST00000119902.6
|
Tspan2
|
tetraspanin 2 |
| chr8_+_34143266 | 0.06 |
ENSMUST00000033992.9
|
Gsr
|
glutathione reductase |
| chr14_+_77394173 | 0.06 |
ENSMUST00000022589.9
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr17_+_37274714 | 0.06 |
ENSMUST00000209623.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr10_-_28862289 | 0.06 |
ENSMUST00000152363.8
ENSMUST00000015663.7 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr5_-_135092904 | 0.05 |
ENSMUST00000111205.8
ENSMUST00000141309.2 |
Bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr10_-_78688000 | 0.05 |
ENSMUST00000205100.3
|
Olfr1356
|
olfactory receptor 1356 |
| chr3_+_126390951 | 0.05 |
ENSMUST00000171289.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr1_-_171854818 | 0.04 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr2_-_44912927 | 0.04 |
ENSMUST00000202432.4
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr13_-_21586858 | 0.03 |
ENSMUST00000117721.8
ENSMUST00000070785.16 ENSMUST00000116433.2 ENSMUST00000223831.2 ENSMUST00000116434.11 ENSMUST00000224820.2 |
Zkscan3
|
zinc finger with KRAB and SCAN domains 3 |
| chr9_+_38119661 | 0.03 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893 |
| chr14_+_65903878 | 0.03 |
ENSMUST00000069226.7
|
Scara5
|
scavenger receptor class A, member 5 |
| chr6_-_112924205 | 0.02 |
ENSMUST00000088373.11
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr12_+_113038376 | 0.02 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chr18_+_37488174 | 0.01 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr5_+_149942140 | 0.01 |
ENSMUST00000065745.10
ENSMUST00000110496.5 ENSMUST00000201612.2 |
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr13_-_40887444 | 0.00 |
ENSMUST00000224665.2
|
Tfap2a
|
transcription factor AP-2, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc13_Hoxd13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.4 | 1.2 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.4 | 1.1 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.3 | 2.3 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 0.9 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.3 | 1.5 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.2 | 1.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 2.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 0.5 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.2 | 2.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.6 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.8 | GO:0045900 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.7 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.5 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.7 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 3.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.6 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 2.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.2 | GO:1902623 | corticospinal neuron axon guidance(GO:0021966) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.4 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.6 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.2 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 1.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.0 | 0.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.7 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 1.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.6 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.0 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.9 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.4 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.2 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.3 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.3 | 3.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 0.9 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.1 | 0.8 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 1.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 2.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 1.5 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.2 | 1.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 0.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.2 | 2.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.8 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 3.2 | GO:0043539 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.5 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 1.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.6 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 0.2 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 2.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 3.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 2.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |