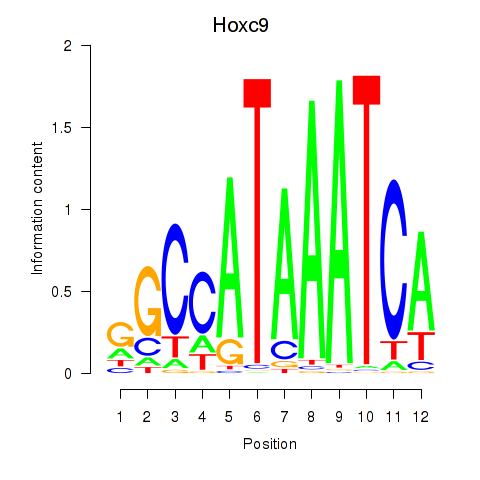
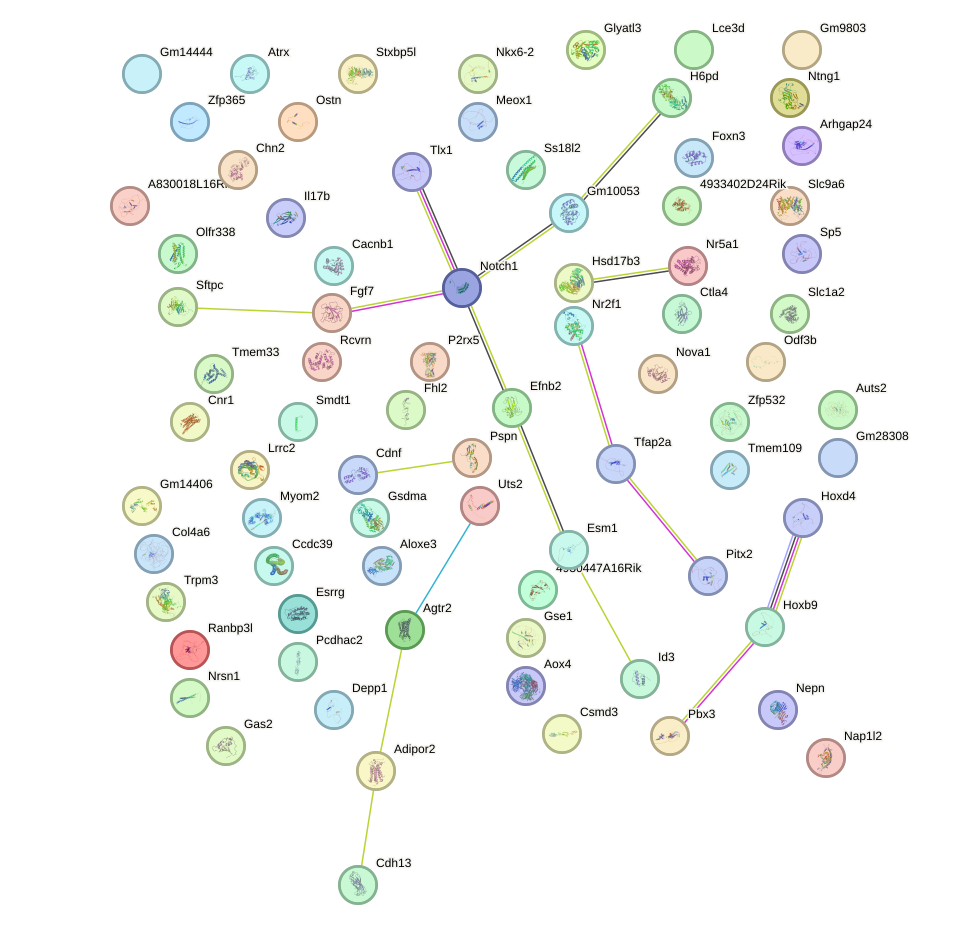
|
chr2_+_102488985
Show fit
|
2.83 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr7_+_51537645
Show fit
|
2.10 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2
|
|
chr4_-_109059414
Show fit
|
1.83 |
ENSMUST00000160774.8
ENSMUST00000194478.6
ENSMUST00000030288.14
ENSMUST00000162787.9
|
Osbpl9
|
oxysterol binding protein-like 9
|
|
chr2_+_70305267
Show fit
|
1.73 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5
|
|
chr7_+_130633776
Show fit
|
1.33 |
ENSMUST00000084509.7
ENSMUST00000213064.3
ENSMUST00000208311.4
|
Dmbt1
|
deleted in malignant brain tumors 1
|
|
chr4_+_135870808
Show fit
|
1.14 |
ENSMUST00000008016.3
|
Id3
|
inhibitor of DNA binding 3
|
|
chr6_-_119365632
Show fit
|
1.07 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2
|
|
chr19_+_24853039
Show fit
|
0.96 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053
|
|
chr17_-_29768531
Show fit
|
0.85 |
ENSMUST00000168339.3
ENSMUST00000114683.10
ENSMUST00000234620.2
|
Tmem217
|
transmembrane protein 217
|
|
chr2_+_3514071
Show fit
|
0.84 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor
|
|
chr10_-_125225298
Show fit
|
0.82 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7
|
|
chr1_+_11484313
Show fit
|
0.80 |
ENSMUST00000171690.9
ENSMUST00000048613.14
|
A830018L16Rik
|
RIKEN cDNA A830018L16 gene
|
|
chr8_+_120955195
Show fit
|
0.76 |
ENSMUST00000180448.3
|
Gse1
|
genetic suppressor element 1, coiled-coil protein
|
|
chr15_-_89263790
Show fit
|
0.76 |
ENSMUST00000238996.2
|
Odf3b
|
outer dense fiber of sperm tails 3B
|
|
chr4_-_91264670
Show fit
|
0.73 |
ENSMUST00000107109.9
ENSMUST00000107111.9
ENSMUST00000107120.8
|
Elavl2
|
ELAV like RNA binding protein 1
|
|
chrX_-_104972150
Show fit
|
0.71 |
ENSMUST00000101305.9
|
Atrx
|
ATRX, chromatin remodeler
|
|
chr1_+_11484489
Show fit
|
0.69 |
ENSMUST00000135014.8
|
A830018L16Rik
|
RIKEN cDNA A830018L16 gene
|
|
chr6_+_54246120
Show fit
|
0.68 |
ENSMUST00000114401.8
|
Chn2
|
chimerin 2
|
|
chr4_-_91264727
Show fit
|
0.67 |
ENSMUST00000107124.10
|
Elavl2
|
ELAV like RNA binding protein 1
|
|
chr5_-_132570710
Show fit
|
0.67 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2
|
|
chr10_-_67748461
Show fit
|
0.62 |
ENSMUST00000064656.8
|
Zfp365
|
zinc finger protein 365
|
|
chr7_-_139162706
Show fit
|
0.61 |
ENSMUST00000106095.3
|
Nkx6-2
|
NK6 homeobox 2
|
|
chr5_+_67417908
Show fit
|
0.58 |
ENSMUST00000037918.12
ENSMUST00000162543.8
ENSMUST00000161233.8
ENSMUST00000160352.8
|
Tmem33
|
transmembrane protein 33
|
|
chr5_+_100054110
Show fit
|
0.58 |
ENSMUST00000198837.3
|
Vamp9
|
vesicle-associated membrane protein 9
|
|
chrX_-_18327610
Show fit
|
0.56 |
ENSMUST00000044188.5
|
Dipk2b
|
divergent protein kinase domain 2B
|
|
chr2_-_34261121
Show fit
|
0.52 |
ENSMUST00000127353.3
ENSMUST00000141653.3
|
Pbx3
|
pre B cell leukemia homeobox 3
|
|
chr4_+_118217179
Show fit
|
0.52 |
ENSMUST00000006562.6
|
Hyi
|
hydroxypyruvate isomerase (putative)
|
|
chr13_-_40891849
Show fit
|
0.48 |
ENSMUST00000223869.2
|
Tfap2a
|
transcription factor AP-2, alpha
|
|
chr2_-_38604503
Show fit
|
0.47 |
ENSMUST00000028084.5
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chr1_+_60948149
Show fit
|
0.45 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr11_+_98555167
Show fit
|
0.43 |
ENSMUST00000017348.3
|
Gsdma
|
gasdermin A
|
|
chr12_-_99529767
Show fit
|
0.42 |
ENSMUST00000176928.3
ENSMUST00000223484.2
|
Foxn3
|
forkhead box N3
|
|
chr6_-_52190299
Show fit
|
0.42 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308
|
|
chr3_-_51184730
Show fit
|
0.41 |
ENSMUST00000195432.2
ENSMUST00000091144.11
ENSMUST00000156983.3
|
Elf2
|
E74-like factor 2
|
|
chr15_+_37425798
Show fit
|
0.40 |
ENSMUST00000022897.2
|
4930447A16Rik
|
RIKEN cDNA 4930447A16 gene
|
|
chr8_+_119010458
Show fit
|
0.39 |
ENSMUST00000117160.2
|
Cdh13
|
cadherin 13
|
|
chr2_+_125876883
Show fit
|
0.39 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7
|
|
chr16_+_27126239
Show fit
|
0.38 |
ENSMUST00000066852.9
|
Ostn
|
osteocrin
|
|
chr18_+_61820982
Show fit
|
0.38 |
ENSMUST00000025471.4
|
Il17b
|
interleukin 17B
|
|
chr1_-_43235914
Show fit
|
0.36 |
ENSMUST00000187357.2
|
Fhl2
|
four and a half LIM domains 2
|
|
chr19_+_22425565
Show fit
|
0.34 |
ENSMUST00000037901.14
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr13_-_25454058
Show fit
|
0.33 |
ENSMUST00000057866.13
|
Nrsn1
|
neurensin 1
|
|
chr3_+_128993609
Show fit
|
0.32 |
ENSMUST00000173645.4
|
Pitx2
|
paired-like homeodomain transcription factor 2
|
|
chr3_-_110050312
Show fit
|
0.32 |
ENSMUST00000156177.9
|
Ntng1
|
netrin G1
|
|
chr3_-_33898405
Show fit
|
0.32 |
ENSMUST00000029222.8
|
Ccdc39
|
coiled-coil domain containing 39
|
|
chr3_+_128993527
Show fit
|
0.32 |
ENSMUST00000174661.9
|
Pitx2
|
paired-like homeodomain transcription factor 2
|
|
chr8_+_15107646
Show fit
|
0.32 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2
|
|
chr19_+_22425534
Show fit
|
0.32 |
ENSMUST00000235522.2
ENSMUST00000236372.2
ENSMUST00000238066.2
ENSMUST00000235780.2
ENSMUST00000236804.2
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr19_-_10857734
Show fit
|
0.31 |
ENSMUST00000133303.8
|
Tmem109
|
transmembrane protein 109
|
|
chr17_+_24645615
Show fit
|
0.29 |
ENSMUST00000234304.2
ENSMUST00000024946.7
ENSMUST00000234150.2
|
Eci1
|
enoyl-Coenzyme A delta isomerase 1
|
|
chr10_+_4660119
Show fit
|
0.28 |
ENSMUST00000105588.8
ENSMUST00000105589.2
|
Esr1
|
estrogen receptor 1 (alpha)
|
|
chr13_-_64237011
Show fit
|
0.28 |
ENSMUST00000166224.8
ENSMUST00000222810.2
|
Hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3
|
|
chr7_+_99384352
Show fit
|
0.27 |
ENSMUST00000098264.2
|
Olfr520
|
olfactory receptor 520
|
|
chr10_+_43400074
Show fit
|
0.27 |
ENSMUST00000057649.8
ENSMUST00000216543.2
|
Gm9803
|
predicted gene 9803
|
|
chr15_+_8997480
Show fit
|
0.26 |
ENSMUST00000227191.3
|
Ranbp3l
|
RAN binding protein 3-like
|
|
chr13_-_64236994
Show fit
|
0.26 |
ENSMUST00000039832.7
|
Hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3
|
|
chr15_+_82230155
Show fit
|
0.26 |
ENSMUST00000023086.15
|
Smdt1
|
single-pass membrane protein with aspartate rich tail 1
|
|
chrX_-_142716085
Show fit
|
0.25 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin
|
|
chr14_+_53315909
Show fit
|
0.25 |
ENSMUST00000103608.4
|
Trav14d-3-dv8
|
T cell receptor alpha variable 14D-3-DV8
|
|
chr11_-_97913420
Show fit
|
0.25 |
ENSMUST00000103144.10
ENSMUST00000017552.13
ENSMUST00000092736.11
ENSMUST00000107562.2
|
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr13_-_78345932
Show fit
|
0.24 |
ENSMUST00000127137.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr18_+_65715351
Show fit
|
0.24 |
ENSMUST00000235669.2
ENSMUST00000183319.8
ENSMUST00000236523.2
ENSMUST00000237886.2
|
Zfp532
|
zinc finger protein 532
|
|
chr11_+_96162283
Show fit
|
0.23 |
ENSMUST00000000010.9
ENSMUST00000174042.3
|
Hoxb9
|
homeobox B9
|
|
chr10_+_52267702
Show fit
|
0.22 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan
|
|
chr16_-_37205302
Show fit
|
0.22 |
ENSMUST00000114781.8
ENSMUST00000114780.8
|
Stxbp5l
|
syntaxin binding protein 5-like
|
|
chr9_+_121539395
Show fit
|
0.22 |
ENSMUST00000035113.11
ENSMUST00000215966.2
ENSMUST00000215833.2
ENSMUST00000215104.2
|
Ss18l2
|
SS18, nBAF chromatin remodeling complex subunit like 2
|
|
chr2_+_125876566
Show fit
|
0.22 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7
|
|
chrX_-_140257030
Show fit
|
0.21 |
ENSMUST00000101205.3
|
Col4a6
|
collagen, type IV, alpha 6
|
|
chr13_-_64237036
Show fit
|
0.21 |
ENSMUST00000222783.2
|
Hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3
|
|
chr6_+_116627567
Show fit
|
0.21 |
ENSMUST00000067354.10
ENSMUST00000178241.4
|
Depp1
|
DEPP1 autophagy regulator
|
|
chr2_-_177269992
Show fit
|
0.21 |
ENSMUST00000108945.8
ENSMUST00000108943.2
|
Gm14406
|
predicted gene 14406
|
|
chr1_+_66360865
Show fit
|
0.21 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2
|
|
chr11_+_85243970
Show fit
|
0.20 |
ENSMUST00000108056.8
ENSMUST00000108061.8
ENSMUST00000108062.8
ENSMUST00000138423.8
ENSMUST00000092821.10
ENSMUST00000074875.11
|
Bcas3
|
breast carcinoma amplified sequence 3
|
|
chr2_+_174852034
Show fit
|
0.20 |
ENSMUST00000109069.8
ENSMUST00000109070.3
|
Gm14444
|
predicted gene 14444
|
|
chr4_+_33924632
Show fit
|
0.20 |
ENSMUST00000057188.7
|
Cnr1
|
cannabinoid receptor 1 (brain)
|
|
chr14_+_54000594
Show fit
|
0.20 |
ENSMUST00000103589.6
|
Trav14-3
|
T cell receptor alpha variable 14-3
|
|
chr1_+_60948307
Show fit
|
0.20 |
ENSMUST00000097720.4
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr3_+_92864693
Show fit
|
0.18 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D
|
|
chr13_+_113346193
Show fit
|
0.18 |
ENSMUST00000038144.9
|
Esm1
|
endothelial cell-specific molecule 1
|
|
chr3_+_128993568
Show fit
|
0.18 |
ENSMUST00000029657.16
ENSMUST00000106382.11
|
Pitx2
|
paired-like homeodomain transcription factor 2
|
|
chr19_+_45139098
Show fit
|
0.18 |
ENSMUST00000026236.11
|
Tlx1
|
T cell leukemia, homeobox 1
|
|
chr11_+_67586140
Show fit
|
0.17 |
ENSMUST00000021290.2
|
Rcvrn
|
recoverin
|
|
chrX_+_21350783
Show fit
|
0.17 |
ENSMUST00000089188.9
|
Agtr2
|
angiotensin II receptor, type 2
|
|
chr17_-_41225241
Show fit
|
0.17 |
ENSMUST00000166343.3
|
Glyatl3
|
glycine-N-acyltransferase-like 3
|
|
chr16_-_37205277
Show fit
|
0.16 |
ENSMUST00000114787.8
ENSMUST00000114782.8
ENSMUST00000114775.8
|
Stxbp5l
|
syntaxin binding protein 5-like
|
|
chr6_+_116627635
Show fit
|
0.16 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator
|
|
chr18_+_37822865
Show fit
|
0.16 |
ENSMUST00000195112.2
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2
|
|
chr9_+_110782861
Show fit
|
0.16 |
ENSMUST00000196834.2
|
Lrrc2
|
leucine rich repeat containing 2
|
|
chr2_+_74552322
Show fit
|
0.15 |
ENSMUST00000047904.4
|
Hoxd4
|
homeobox D4
|
|
chrX_+_55655111
Show fit
|
0.15 |
ENSMUST00000144068.8
ENSMUST00000077741.12
ENSMUST00000114784.4
|
Slc9a6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr6_-_55109960
Show fit
|
0.14 |
ENSMUST00000003568.15
|
Crhr2
|
corticotropin releasing hormone receptor 2
|
|
chr2_-_26406631
Show fit
|
0.12 |
ENSMUST00000132820.2
|
Notch1
|
notch 1
|
|
chr17_-_57307018
Show fit
|
0.11 |
ENSMUST00000002740.3
|
Pspn
|
persephin
|
|
chrX_-_142716200
Show fit
|
0.10 |
ENSMUST00000112851.8
ENSMUST00000112856.3
ENSMUST00000033642.10
|
Dcx
|
doublecortin
|
|
chr11_+_73051228
Show fit
|
0.10 |
ENSMUST00000006104.10
ENSMUST00000135202.8
ENSMUST00000136894.3
|
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr11_-_101785181
Show fit
|
0.09 |
ENSMUST00000057054.8
|
Meox1
|
mesenchyme homeobox 1
|
|
chr13_-_96607619
Show fit
|
0.08 |
ENSMUST00000239026.2
ENSMUST00000181613.3
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B
|
|
chr1_+_187730018
Show fit
|
0.07 |
ENSMUST00000027906.13
|
Esrrg
|
estrogen-related receptor gamma
|
|
chr15_-_48655329
Show fit
|
0.07 |
ENSMUST00000160658.8
ENSMUST00000100670.10
ENSMUST00000162830.8
|
Csmd3
|
CUB and Sushi multiple domains 3
|
|
chr10_+_69761784
Show fit
|
0.06 |
ENSMUST00000181974.8
ENSMUST00000182795.8
ENSMUST00000182437.8
|
Ank3
|
ankyrin 3, epithelial
|
|
chr4_-_150087587
Show fit
|
0.06 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase)
|
|
chr5_+_102916637
Show fit
|
0.06 |
ENSMUST00000112852.8
|
Arhgap24
|
Rho GTPase activating protein 24
|
|
chr12_-_46767619
Show fit
|
0.05 |
ENSMUST00000219886.2
|
Nova1
|
NOVA alternative splicing regulator 1
|
|
chr1_-_63808426
Show fit
|
0.05 |
ENSMUST00000027102.2
|
4933402D24Rik
|
RIKEN cDNA 4933402D24 gene
|
|
chr6_+_134373275
Show fit
|
0.04 |
ENSMUST00000032321.11
|
Bcl2l14
|
BCL2-like 14 (apoptosis facilitator)
|
|
chr1_+_58249556
Show fit
|
0.04 |
ENSMUST00000040442.6
|
Aox4
|
aldehyde oxidase 4
|
|
chr13_-_96607722
Show fit
|
0.04 |
ENSMUST00000055607.13
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B
|
|
chr1_+_187730032
Show fit
|
0.02 |
ENSMUST00000110938.2
|
Esrrg
|
estrogen-related receptor gamma
|
|
chr11_+_69016722
Show fit
|
0.02 |
ENSMUST00000021268.9
|
Aloxe3
|
arachidonate lipoxygenase 3
|
|
chr3_-_51184895
Show fit
|
0.01 |
ENSMUST00000108051.8
ENSMUST00000108053.9
|
Elf2
|
E74-like factor 2
|
|
chrX_-_102230225
Show fit
|
0.01 |
ENSMUST00000121720.2
|
Nap1l2
|
nucleosome assembly protein 1-like 2
|
|
chr4_+_151081538
Show fit
|
0.00 |
ENSMUST00000030803.2
|
Uts2
|
urotensin 2
|