Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
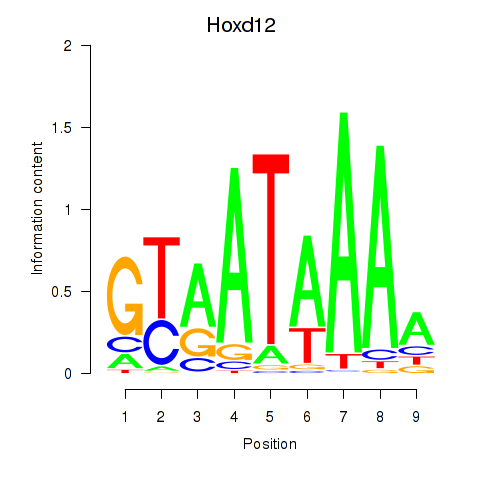
Results for Hoxd12
Z-value: 0.64
Transcription factors associated with Hoxd12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd12
|
ENSMUSG00000001823.6 | homeobox D12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd12 | mm39_v1_chr2_+_74505350_74505357 | -0.27 | 1.1e-01 | Click! |
Activity profile of Hoxd12 motif
Sorted Z-values of Hoxd12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_138121245 | 4.03 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr19_+_40078132 | 3.93 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr19_+_24853039 | 2.07 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr8_-_5155347 | 1.66 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chrM_+_5319 | 1.35 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr8_-_70355208 | 1.08 |
ENSMUST00000110167.5
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr7_+_123061535 | 1.05 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr7_+_123061497 | 1.01 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr2_+_155453103 | 0.97 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr6_-_119365632 | 0.89 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr10_-_30531768 | 0.86 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr4_+_43641262 | 0.86 |
ENSMUST00000123351.8
ENSMUST00000128549.3 |
Npr2
|
natriuretic peptide receptor 2 |
| chr17_+_3447465 | 0.83 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr16_+_42727926 | 0.78 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_-_30531832 | 0.76 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr17_+_31652029 | 0.73 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr16_-_35589726 | 0.73 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr1_-_57008986 | 0.73 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr16_+_11223512 | 0.70 |
ENSMUST00000096273.9
|
Snx29
|
sorting nexin 29 |
| chr17_+_31652073 | 0.70 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr1_-_46927230 | 0.69 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr19_-_45800730 | 0.69 |
ENSMUST00000086993.11
|
Kcnip2
|
Kv channel-interacting protein 2 |
| chr4_+_31964082 | 0.68 |
ENSMUST00000037607.11
ENSMUST00000080933.13 ENSMUST00000108183.8 ENSMUST00000108184.3 |
Map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr3_-_154036180 | 0.63 |
ENSMUST00000177846.8
|
Lhx8
|
LIM homeobox protein 8 |
| chr13_-_23806530 | 0.63 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr14_-_68771138 | 0.62 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr10_+_43400074 | 0.60 |
ENSMUST00000057649.8
ENSMUST00000216543.2 |
Gm9803
|
predicted gene 9803 |
| chr3_+_66893280 | 0.58 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr5_+_128677863 | 0.58 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10 |
| chr5_-_101812862 | 0.57 |
ENSMUST00000044125.11
|
Nkx6-1
|
NK6 homeobox 1 |
| chr19_-_50667079 | 0.57 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chrX_+_139357362 | 0.57 |
ENSMUST00000033809.4
|
Prps1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr3_+_75981577 | 0.56 |
ENSMUST00000038364.15
|
Fstl5
|
follistatin-like 5 |
| chr1_+_46464625 | 0.56 |
ENSMUST00000189749.7
|
Dnah7c
|
dynein, axonemal, heavy chain 7C |
| chr3_+_103482547 | 0.56 |
ENSMUST00000121834.8
|
Syt6
|
synaptotagmin VI |
| chr3_+_103482591 | 0.56 |
ENSMUST00000090697.11
ENSMUST00000239027.2 |
Syt6
|
synaptotagmin VI |
| chrM_+_3906 | 0.55 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr7_+_43339842 | 0.53 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr8_+_45960804 | 0.52 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr7_+_80863314 | 0.51 |
ENSMUST00000026672.8
|
Pde8a
|
phosphodiesterase 8A |
| chr6_+_71176811 | 0.50 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr1_-_176041498 | 0.50 |
ENSMUST00000111167.2
|
Pld5
|
phospholipase D family, member 5 |
| chr8_+_45960931 | 0.49 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr14_-_48900192 | 0.48 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr11_+_31823096 | 0.48 |
ENSMUST00000155278.2
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr10_-_116732813 | 0.47 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr7_+_89780785 | 0.47 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr10_+_34359395 | 0.46 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr18_-_3281727 | 0.46 |
ENSMUST00000154705.8
ENSMUST00000151084.8 |
Crem
|
cAMP responsive element modulator |
| chrX_-_142716085 | 0.46 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr10_-_42459624 | 0.45 |
ENSMUST00000019938.11
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr17_-_36207965 | 0.44 |
ENSMUST00000150056.2
ENSMUST00000156817.2 ENSMUST00000146451.8 ENSMUST00000148482.8 |
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr10_+_23814470 | 0.44 |
ENSMUST00000079134.5
|
Taar2
|
trace amine-associated receptor 2 |
| chr8_-_47805383 | 0.44 |
ENSMUST00000110367.10
|
Stox2
|
storkhead box 2 |
| chr10_+_52267702 | 0.44 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr15_+_8997480 | 0.44 |
ENSMUST00000227191.3
|
Ranbp3l
|
RAN binding protein 3-like |
| chr3_-_141637245 | 0.43 |
ENSMUST00000106232.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr3_+_66893031 | 0.43 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr9_-_89620461 | 0.42 |
ENSMUST00000060700.4
ENSMUST00000185470.3 |
Ankrd34c
|
ankyrin repeat domain 34C |
| chr17_-_91400142 | 0.42 |
ENSMUST00000160800.9
|
Nrxn1
|
neurexin I |
| chr8_-_25528972 | 0.41 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr10_-_67972401 | 0.40 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr2_+_57887896 | 0.40 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr1_+_128069716 | 0.39 |
ENSMUST00000187557.2
|
R3hdm1
|
R3H domain containing 1 |
| chr13_-_21826688 | 0.39 |
ENSMUST00000205631.3
|
Olfr11
|
olfactory receptor 11 |
| chr10_-_25076008 | 0.39 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr19_+_22425565 | 0.39 |
ENSMUST00000037901.14
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr10_+_4561974 | 0.39 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr9_+_45230370 | 0.38 |
ENSMUST00000034597.8
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr17_-_52133594 | 0.38 |
ENSMUST00000129667.8
ENSMUST00000169480.8 ENSMUST00000148559.2 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr4_-_83203388 | 0.38 |
ENSMUST00000150522.8
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr1_-_165762469 | 0.37 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr5_+_142678212 | 0.36 |
ENSMUST00000198728.5
|
Slc29a4
|
solute carrier family 29 (nucleoside transporters), member 4 |
| chr18_-_20135139 | 0.35 |
ENSMUST00000115848.5
|
Dsc3
|
desmocollin 3 |
| chr18_-_20135012 | 0.35 |
ENSMUST00000225110.2
|
Dsc3
|
desmocollin 3 |
| chr10_+_34359513 | 0.35 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr2_-_103114105 | 0.35 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chr7_+_5017414 | 0.34 |
ENSMUST00000207901.2
|
Zfp524
|
zinc finger protein 524 |
| chrX_-_142716200 | 0.34 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr13_-_43632368 | 0.34 |
ENSMUST00000222651.2
|
Ranbp9
|
RAN binding protein 9 |
| chr11_-_46581135 | 0.34 |
ENSMUST00000169584.8
|
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr5_-_25047577 | 0.34 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr1_+_132128701 | 0.34 |
ENSMUST00000189062.2
|
Lemd1
|
LEM domain containing 1 |
| chr17_-_36208265 | 0.33 |
ENSMUST00000148721.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr10_+_116013256 | 0.33 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr18_-_20135396 | 0.33 |
ENSMUST00000223946.2
|
Dsc3
|
desmocollin 3 |
| chr4_-_154721288 | 0.33 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr10_+_116013122 | 0.33 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr5_-_65855511 | 0.33 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr11_-_88608958 | 0.32 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr3_+_66892979 | 0.32 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chrX_+_35459557 | 0.32 |
ENSMUST00000115258.9
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr14_-_52011035 | 0.32 |
ENSMUST00000073860.6
|
Ang4
|
angiogenin, ribonuclease A family, member 4 |
| chr16_-_89305201 | 0.32 |
ENSMUST00000056118.4
|
Krtap7-1
|
keratin associated protein 7-1 |
| chr17_+_35235552 | 0.31 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr14_-_118289557 | 0.31 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr7_+_30264835 | 0.31 |
ENSMUST00000043850.14
|
Igflr1
|
IGF-like family receptor 1 |
| chr11_-_42070517 | 0.31 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr9_+_40092216 | 0.31 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chrX_+_35459621 | 0.31 |
ENSMUST00000115256.2
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chrX_+_35459589 | 0.30 |
ENSMUST00000048067.10
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr15_+_25774070 | 0.30 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr1_-_69147185 | 0.30 |
ENSMUST00000121473.8
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr1_+_163875783 | 0.30 |
ENSMUST00000027874.6
|
Sele
|
selectin, endothelial cell |
| chr15_-_8739893 | 0.29 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chrX_+_35459601 | 0.29 |
ENSMUST00000115257.8
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr13_-_53627110 | 0.29 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr5_-_148489593 | 0.29 |
ENSMUST00000201595.4
ENSMUST00000164904.2 |
Ubl3
|
ubiquitin-like 3 |
| chr9_-_14815050 | 0.28 |
ENSMUST00000148155.2
ENSMUST00000121116.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr14_-_67106037 | 0.28 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr14_+_31807760 | 0.28 |
ENSMUST00000170600.8
ENSMUST00000168986.7 ENSMUST00000169649.2 |
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr10_-_80257681 | 0.27 |
ENSMUST00000156244.2
|
Tcf3
|
transcription factor 3 |
| chrX_-_71961890 | 0.27 |
ENSMUST00000152200.2
|
Cetn2
|
centrin 2 |
| chr17_+_72088281 | 0.27 |
ENSMUST00000229874.2
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr1_+_6805048 | 0.27 |
ENSMUST00000139838.8
|
St18
|
suppression of tumorigenicity 18 |
| chr9_+_65268304 | 0.27 |
ENSMUST00000147185.3
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr12_+_33197776 | 0.27 |
ENSMUST00000125192.9
ENSMUST00000136644.8 |
Atxn7l1
|
ataxin 7-like 1 |
| chr11_-_99996452 | 0.27 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr1_+_128069677 | 0.27 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr16_-_56616186 | 0.26 |
ENSMUST00000023437.5
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr16_+_57173632 | 0.26 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr1_+_82817794 | 0.26 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr19_+_26725589 | 0.26 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_-_43492367 | 0.26 |
ENSMUST00000020672.5
|
Fabp6
|
fatty acid binding protein 6 |
| chr9_-_14815163 | 0.26 |
ENSMUST00000069408.10
|
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr19_+_60744385 | 0.25 |
ENSMUST00000088237.6
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chrX_-_103244728 | 0.25 |
ENSMUST00000056502.7
|
Nexmif
|
neurite extension and migration factor |
| chr11_+_29413734 | 0.25 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr9_-_123778334 | 0.24 |
ENSMUST00000071404.5
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr15_-_34356567 | 0.24 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr2_+_48839505 | 0.24 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr3_-_26386609 | 0.24 |
ENSMUST00000193603.6
|
Nlgn1
|
neuroligin 1 |
| chr10_+_19232281 | 0.23 |
ENSMUST00000053225.7
|
Olig3
|
oligodendrocyte transcription factor 3 |
| chr14_-_31807552 | 0.23 |
ENSMUST00000022461.11
ENSMUST00000067955.12 ENSMUST00000124303.9 |
Dph3
|
diphthamine biosynthesis 3 |
| chr12_+_33197692 | 0.23 |
ENSMUST00000077456.13
ENSMUST00000110824.9 |
Atxn7l1
|
ataxin 7-like 1 |
| chrX_-_103244784 | 0.23 |
ENSMUST00000118314.8
|
Nexmif
|
neurite extension and migration factor |
| chr1_-_37535170 | 0.23 |
ENSMUST00000148047.2
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr12_+_33197753 | 0.22 |
ENSMUST00000146040.9
|
Atxn7l1
|
ataxin 7-like 1 |
| chr14_-_36641270 | 0.22 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr5_-_74692327 | 0.22 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr1_+_173986288 | 0.22 |
ENSMUST00000068403.4
|
Olfr420
|
olfactory receptor 420 |
| chr14_-_36641470 | 0.21 |
ENSMUST00000182042.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr18_-_3281752 | 0.21 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr1_-_37535202 | 0.21 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr18_-_35348049 | 0.21 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr4_-_82423985 | 0.20 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr3_-_19217174 | 0.20 |
ENSMUST00000029125.10
|
Armc1
|
armadillo repeat containing 1 |
| chr17_-_45047521 | 0.20 |
ENSMUST00000113572.9
|
Runx2
|
runt related transcription factor 2 |
| chr10_+_119828821 | 0.20 |
ENSMUST00000105261.9
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr6_+_15185202 | 0.20 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr11_-_101785181 | 0.20 |
ENSMUST00000057054.8
|
Meox1
|
mesenchyme homeobox 1 |
| chr5_-_28672091 | 0.19 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chr3_-_64049349 | 0.19 |
ENSMUST00000177151.9
|
Vmn2r2
|
vomeronasal 2, receptor 2 |
| chr9_-_77255171 | 0.19 |
ENSMUST00000185039.8
|
Mlip
|
muscular LMNA-interacting protein |
| chr2_-_35994819 | 0.19 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr4_+_148085179 | 0.19 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr10_-_107321938 | 0.18 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chrX_+_159551009 | 0.18 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr9_+_109641348 | 0.17 |
ENSMUST00000200555.5
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr16_-_50252703 | 0.17 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr2_-_18053595 | 0.16 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr12_+_55883101 | 0.16 |
ENSMUST00000059250.8
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr7_-_102408573 | 0.16 |
ENSMUST00000210453.3
ENSMUST00000232246.3 ENSMUST00000239110.2 ENSMUST00000060187.15 ENSMUST00000168007.3 |
Olfr560
Olfr78
|
olfactory receptor 560 olfactory receptor 78 |
| chr6_+_15185562 | 0.16 |
ENSMUST00000137628.8
|
Foxp2
|
forkhead box P2 |
| chrX_-_49108236 | 0.16 |
ENSMUST00000213556.3
ENSMUST00000213463.2 |
Olfr1323
|
olfactory receptor 1323 |
| chr17_-_47169380 | 0.16 |
ENSMUST00000233455.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr4_-_82423944 | 0.16 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr8_+_45960855 | 0.16 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr3_-_117153802 | 0.15 |
ENSMUST00000197743.2
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr15_+_81821112 | 0.15 |
ENSMUST00000135663.2
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr2_+_85822163 | 0.15 |
ENSMUST00000050942.3
|
Olfr1031
|
olfactory receptor 1031 |
| chr8_+_70354828 | 0.15 |
ENSMUST00000050373.7
|
Tssk6
|
testis-specific serine kinase 6 |
| chr10_+_87926932 | 0.14 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr2_+_111327525 | 0.14 |
ENSMUST00000121345.4
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr5_+_18167547 | 0.14 |
ENSMUST00000030561.9
|
Gnat3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr13_-_3995349 | 0.14 |
ENSMUST00000058610.8
|
Ucn3
|
urocortin 3 |
| chr18_-_84854841 | 0.14 |
ENSMUST00000236689.2
|
Gm17266
|
predicted gene, 17266 |
| chr6_+_90179768 | 0.14 |
ENSMUST00000078371.6
|
V1ra8
|
vomeronasal 1 receptor, A8 |
| chr1_+_59296065 | 0.13 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr15_+_36179676 | 0.13 |
ENSMUST00000171205.3
|
Spag1
|
sperm associated antigen 1 |
| chr9_+_43222104 | 0.13 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr2_+_73102269 | 0.13 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
| chr4_-_82423511 | 0.12 |
ENSMUST00000050872.15
ENSMUST00000064770.9 |
Nfib
|
nuclear factor I/B |
| chr12_-_69838004 | 0.11 |
ENSMUST00000221646.2
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr7_+_103379581 | 0.11 |
ENSMUST00000098193.4
|
Olfr628
|
olfactory receptor 628 |
| chr15_-_8740218 | 0.11 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr14_+_26616514 | 0.11 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr14_+_53048391 | 0.11 |
ENSMUST00000103646.5
|
Trav10d
|
T cell receptor alpha variable 10D |
| chr2_-_45007407 | 0.10 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_143864993 | 0.10 |
ENSMUST00000172367.3
|
Gm42421
|
predicted gene, 42421 |
| chr6_+_136528155 | 0.10 |
ENSMUST00000186742.2
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr6_+_15185399 | 0.09 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr2_+_65676176 | 0.09 |
ENSMUST00000053910.10
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chrX_+_159551171 | 0.09 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr13_+_83672389 | 0.09 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr9_+_110948492 | 0.09 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr9_-_123507937 | 0.08 |
ENSMUST00000040960.13
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr6_-_38230890 | 0.08 |
ENSMUST00000117556.8
ENSMUST00000169256.5 |
D630045J12Rik
|
RIKEN cDNA D630045J12 gene |
| chr9_-_14815228 | 0.08 |
ENSMUST00000034409.14
ENSMUST00000117620.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr4_+_136012019 | 0.08 |
ENSMUST00000130223.8
|
Zfp46
|
zinc finger protein 46 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 3.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.3 | 2.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 1.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.2 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 0.6 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 | 1.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.7 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 1.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.3 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 0.3 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.6 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.6 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.7 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 1.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.5 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.5 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.2 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.4 | GO:0021960 | amygdala development(GO:0021764) anterior commissure morphogenesis(GO:0021960) regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.3 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 0.8 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.6 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.4 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.1 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.3 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 1.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.7 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.2 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.5 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 1.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 1.2 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.5 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.0 | GO:1904499 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.4 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.3 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.0 | 3.9 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.2 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.9 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 0.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.4 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 2.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.3 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 1.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |