Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
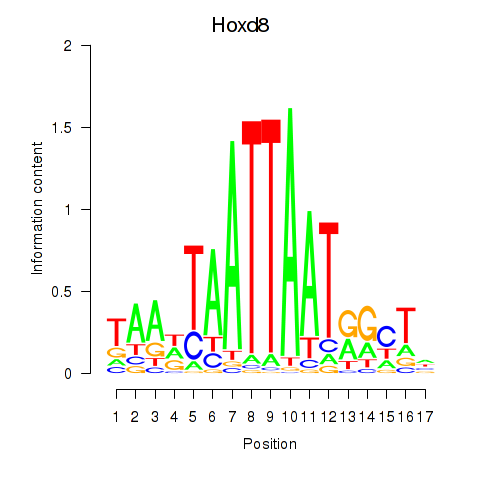
Results for Hoxd8
Z-value: 0.79
Transcription factors associated with Hoxd8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd8
|
ENSMUSG00000027102.5 | homeobox D8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd8 | mm39_v1_chr2_+_74535489_74535500 | -0.23 | 1.8e-01 | Click! |
Activity profile of Hoxd8 motif
Sorted Z-values of Hoxd8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_121983720 | 9.00 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chr19_-_46661321 | 4.41 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr7_+_140343652 | 4.24 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr19_-_46661501 | 4.12 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr10_-_127843377 | 3.49 |
ENSMUST00000219447.2
ENSMUST00000219780.2 ENSMUST00000219707.2 ENSMUST00000219953.2 ENSMUST00000219183.2 |
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr18_-_38999755 | 3.35 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr1_+_88093726 | 3.06 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr5_+_87148697 | 2.93 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr19_-_39637489 | 2.75 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr1_+_9618173 | 2.66 |
ENSMUST00000144177.8
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr9_+_80072274 | 2.59 |
ENSMUST00000035889.15
ENSMUST00000113268.8 |
Myo6
|
myosin VI |
| chr14_+_55797443 | 2.46 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr3_+_94280101 | 2.38 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr9_+_80072361 | 2.11 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr1_+_180878797 | 2.09 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr14_+_55797468 | 2.07 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr18_-_39000056 | 1.92 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr15_-_96929086 | 1.88 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr9_-_15212849 | 1.84 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr11_+_117700479 | 1.82 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr13_+_24023428 | 1.76 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr13_-_63036096 | 1.71 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr7_-_12829100 | 1.67 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr3_+_142300601 | 1.66 |
ENSMUST00000029936.5
|
Gbp2b
|
guanylate binding protein 2b |
| chr1_+_87983099 | 1.54 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr17_+_79919267 | 1.41 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr13_+_24023386 | 1.35 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr2_+_152511381 | 1.33 |
ENSMUST00000125366.8
ENSMUST00000109825.8 ENSMUST00000089059.9 ENSMUST00000079247.4 |
H13
|
histocompatibility 13 |
| chr1_+_87983189 | 1.22 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr9_+_21634779 | 1.21 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr7_-_44711771 | 1.14 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr7_-_99629637 | 1.09 |
ENSMUST00000080817.6
|
Rnf169
|
ring finger protein 169 |
| chr17_+_44114894 | 1.02 |
ENSMUST00000044895.13
|
Rcan2
|
regulator of calcineurin 2 |
| chr3_-_75177378 | 1.02 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr12_-_25147139 | 0.93 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr3_+_20011405 | 0.93 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr11_-_59466995 | 0.91 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr1_-_138776315 | 0.90 |
ENSMUST00000112030.9
|
Lhx9
|
LIM homeobox protein 9 |
| chr5_+_29400981 | 0.90 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr7_+_130633776 | 0.85 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr2_-_86208737 | 0.85 |
ENSMUST00000217435.2
|
Olfr1057
|
olfactory receptor 1057 |
| chr4_-_108158242 | 0.80 |
ENSMUST00000043616.7
|
Zyg11b
|
zyg-ll family member B, cell cycle regulator |
| chr6_+_137387729 | 0.80 |
ENSMUST00000203914.2
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr17_+_46807637 | 0.78 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr16_+_11224481 | 0.75 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr11_+_77928736 | 0.74 |
ENSMUST00000072289.12
ENSMUST00000100784.9 ENSMUST00000073660.7 |
Flot2
|
flotillin 2 |
| chr3_-_19319123 | 0.73 |
ENSMUST00000121951.2
|
Pde7a
|
phosphodiesterase 7A |
| chr17_+_34866160 | 0.68 |
ENSMUST00000173984.2
|
Atf6b
|
activating transcription factor 6 beta |
| chr9_-_55419442 | 0.63 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr15_-_103473481 | 0.61 |
ENSMUST00000228060.2
ENSMUST00000228895.2 ENSMUST00000023134.5 |
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr2_+_81883566 | 0.60 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr2_-_163314480 | 0.59 |
ENSMUST00000109418.2
|
Fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr7_+_43077088 | 0.58 |
ENSMUST00000239023.2
|
Gm38999
|
predicted gene, 38999 |
| chr2_+_151336197 | 0.58 |
ENSMUST00000028949.16
ENSMUST00000103160.5 |
Nsfl1c
|
NSFL1 (p97) cofactor (p47) |
| chr1_+_178233640 | 0.57 |
ENSMUST00000027775.9
|
Efcab2
|
EF-hand calcium binding domain 2 |
| chr10_+_103203552 | 0.57 |
ENSMUST00000179636.3
ENSMUST00000217905.2 ENSMUST00000074204.12 |
Slc6a15
|
solute carrier family 6 (neurotransmitter transporter), member 15 |
| chr5_-_38649291 | 0.56 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr7_-_80055168 | 0.56 |
ENSMUST00000107362.10
ENSMUST00000135306.3 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr12_-_84664001 | 0.55 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr18_-_73887528 | 0.55 |
ENSMUST00000041138.3
|
Elac1
|
elaC ribonuclease Z 1 |
| chr17_+_34866090 | 0.54 |
ENSMUST00000015605.15
|
Atf6b
|
activating transcription factor 6 beta |
| chr12_-_99529767 | 0.53 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr15_+_99192968 | 0.53 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr6_+_134617903 | 0.50 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chrX_-_158921370 | 0.49 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr11_+_109434519 | 0.46 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr8_+_46463633 | 0.45 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr3_-_96104886 | 0.45 |
ENSMUST00000016087.4
|
Bola1
|
bolA-like 1 (E. coli) |
| chr10_+_116013256 | 0.44 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr10_+_23770586 | 0.44 |
ENSMUST00000041416.8
|
Vnn1
|
vanin 1 |
| chr9_-_82856321 | 0.44 |
ENSMUST00000189985.2
|
Phip
|
pleckstrin homology domain interacting protein |
| chr1_-_14380032 | 0.44 |
ENSMUST00000187790.2
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr3_-_95778679 | 0.41 |
ENSMUST00000142437.2
ENSMUST00000067298.5 |
Mrps21
|
mitochondrial ribosomal protein S21 |
| chr11_-_99979052 | 0.41 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr2_+_83554770 | 0.41 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr8_-_117809188 | 0.40 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr1_+_88234454 | 0.40 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr10_+_116013122 | 0.39 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr6_+_30610973 | 0.38 |
ENSMUST00000062758.11
|
Cpa5
|
carboxypeptidase A5 |
| chr17_-_8366536 | 0.38 |
ENSMUST00000231927.2
|
Rnaset2a
|
ribonuclease T2A |
| chr9_+_54446268 | 0.37 |
ENSMUST00000060242.12
|
Sh2d7
|
SH2 domain containing 7 |
| chr9_+_72714156 | 0.37 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chrX_-_125723491 | 0.37 |
ENSMUST00000081074.5
|
4932411N23Rik
|
RIKEN cDNA 4932411N23 gene |
| chr19_+_44481901 | 0.37 |
ENSMUST00000041163.5
|
Wnt8b
|
wingless-type MMTV integration site family, member 8B |
| chr19_-_11833365 | 0.37 |
ENSMUST00000079875.4
|
Olfr1418
|
olfactory receptor 1418 |
| chr2_+_87854404 | 0.36 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chrX_+_118836893 | 0.35 |
ENSMUST00000040961.3
ENSMUST00000113366.2 |
Pabpc5
|
poly(A) binding protein, cytoplasmic 5 |
| chr6_+_30611028 | 0.35 |
ENSMUST00000115138.8
|
Cpa5
|
carboxypeptidase A5 |
| chr1_+_177557380 | 0.35 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr7_-_29605949 | 0.35 |
ENSMUST00000159920.2
ENSMUST00000162592.8 |
Zfp27
|
zinc finger protein 27 |
| chr1_+_178626003 | 0.34 |
ENSMUST00000160789.2
|
Kif26b
|
kinesin family member 26B |
| chr2_+_87260619 | 0.33 |
ENSMUST00000215909.2
|
Olfr1124
|
olfactory receptor 1124 |
| chr2_-_151510453 | 0.33 |
ENSMUST00000180195.8
ENSMUST00000096439.4 |
Rad21l
|
RAD21-like (S. pombe) |
| chr12_+_87490666 | 0.32 |
ENSMUST00000161023.8
ENSMUST00000160488.8 ENSMUST00000077462.8 ENSMUST00000160880.2 |
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr10_+_34265969 | 0.32 |
ENSMUST00000105511.2
|
Col10a1
|
collagen, type X, alpha 1 |
| chr6_+_123755838 | 0.31 |
ENSMUST00000075095.7
ENSMUST00000233759.2 ENSMUST00000232985.2 |
Vmn2r24
|
vomeronasal 2, receptor 24 |
| chr4_+_118818775 | 0.31 |
ENSMUST00000058651.5
|
Lao1
|
L-amino acid oxidase 1 |
| chr8_-_85389470 | 0.29 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr9_-_99302205 | 0.28 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr2_-_88909981 | 0.28 |
ENSMUST00000213724.2
|
Olfr1219
|
olfactory receptor 1219 |
| chr6_-_129077867 | 0.28 |
ENSMUST00000032258.8
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr13_+_38388904 | 0.28 |
ENSMUST00000091641.13
ENSMUST00000178564.2 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr1_-_160405464 | 0.28 |
ENSMUST00000238289.2
|
Gpr52
|
G protein-coupled receptor 52 |
| chr2_+_89757653 | 0.28 |
ENSMUST00000213720.3
ENSMUST00000102609.3 |
Olfr1258
|
olfactory receptor 1258 |
| chr3_-_30194559 | 0.27 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chrM_+_7779 | 0.27 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr5_-_121665249 | 0.27 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr6_-_122443494 | 0.25 |
ENSMUST00000204731.2
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr14_-_50521663 | 0.25 |
ENSMUST00000213701.2
|
Olfr732
|
olfactory receptor 732 |
| chr11_-_87783073 | 0.25 |
ENSMUST00000213672.2
ENSMUST00000213928.2 |
Olfr462
|
olfactory receptor 462 |
| chrX_+_158242121 | 0.25 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr8_-_22396428 | 0.25 |
ENSMUST00000051965.5
|
Defb11
|
defensin beta 11 |
| chr17_+_37977879 | 0.25 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr9_-_40015750 | 0.25 |
ENSMUST00000213858.2
|
Olfr984
|
olfactory receptor 984 |
| chr1_+_65321215 | 0.23 |
ENSMUST00000140190.8
|
Pth2r
|
parathyroid hormone 2 receptor |
| chr7_+_17979272 | 0.23 |
ENSMUST00000066780.5
|
Mill1
|
MHC I like leukocyte 1 |
| chr4_+_88768324 | 0.23 |
ENSMUST00000094972.2
|
Ifna1
|
interferon alpha 1 |
| chr14_-_20443676 | 0.22 |
ENSMUST00000061444.5
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chrY_+_55729767 | 0.22 |
ENSMUST00000177834.2
|
Gm21858
|
predicted gene, 21858 |
| chr2_-_89678487 | 0.22 |
ENSMUST00000214428.3
|
Olfr48
|
olfactory receptor 48 |
| chr10_-_108535970 | 0.21 |
ENSMUST00000218023.2
|
Gm5136
|
predicted gene 5136 |
| chr7_-_103191732 | 0.21 |
ENSMUST00000215663.2
|
Olfr612
|
olfactory receptor 612 |
| chr1_-_63153675 | 0.21 |
ENSMUST00000097718.9
|
Ino80d
|
INO80 complex subunit D |
| chr3_-_96359622 | 0.21 |
ENSMUST00000093126.11
ENSMUST00000098841.4 |
BC107364
|
cDNA sequence BC107364 |
| chr4_+_101843823 | 0.21 |
ENSMUST00000106914.8
|
Gm12789
|
predicted gene 12789 |
| chr4_+_148985584 | 0.20 |
ENSMUST00000147270.2
|
Casz1
|
castor zinc finger 1 |
| chrY_-_35875442 | 0.20 |
ENSMUST00000180332.2
|
Gm20896
|
predicted gene, 20896 |
| chrY_+_84109980 | 0.20 |
ENSMUST00000177775.2
|
Gm21095
|
predicted gene, 21095 |
| chr5_-_66672158 | 0.20 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr8_-_65440483 | 0.19 |
ENSMUST00000210982.2
|
Trim75
|
tripartite motif-containing 75 |
| chr1_-_134477400 | 0.18 |
ENSMUST00000172898.2
|
Mgat4e
|
MGAT4 family, member E |
| chr8_-_65440298 | 0.18 |
ENSMUST00000095295.3
|
Trim75
|
tripartite motif-containing 75 |
| chrY_+_55211732 | 0.18 |
ENSMUST00000180249.2
|
Gm20931
|
predicted gene, 20931 |
| chr10_+_116881246 | 0.18 |
ENSMUST00000073834.5
|
Lrrc10
|
leucine rich repeat containing 10 |
| chr7_-_103394150 | 0.17 |
ENSMUST00000213906.2
|
Olfr629
|
olfactory receptor 629 |
| chr2_+_83554868 | 0.17 |
ENSMUST00000111740.9
|
Itgav
|
integrin alpha V |
| chrY_+_65387652 | 0.16 |
ENSMUST00000178198.2
|
Gm20736
|
predicted gene, 20736 |
| chr4_+_109092829 | 0.16 |
ENSMUST00000030285.8
|
Calr4
|
calreticulin 4 |
| chrY_+_79332266 | 0.16 |
ENSMUST00000178063.2
|
Gm20916
|
predicted gene, 20916 |
| chr10_+_129403524 | 0.16 |
ENSMUST00000204820.3
|
Olfr794
|
olfactory receptor 794 |
| chr2_+_83554741 | 0.16 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr3_-_144555062 | 0.15 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chrM_+_7758 | 0.15 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr2_-_165210622 | 0.15 |
ENSMUST00000141140.2
ENSMUST00000103085.8 |
Zfp663
|
zinc finger protein 663 |
| chr19_-_12313274 | 0.15 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr3_-_33136153 | 0.15 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chrY_+_52778041 | 0.14 |
ENSMUST00000178673.2
|
Gm21258
|
predicted gene, 21258 |
| chr8_+_22329942 | 0.14 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chr6_-_69920632 | 0.13 |
ENSMUST00000198880.5
ENSMUST00000103371.3 |
Igkv12-38
|
immunoglobulin kappa chain variable 12-38 |
| chr11_+_73489420 | 0.13 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr2_-_73410632 | 0.13 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr1_+_58841808 | 0.12 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr10_-_41570483 | 0.12 |
ENSMUST00000190522.2
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr19_-_18978463 | 0.12 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr11_+_16207705 | 0.12 |
ENSMUST00000109645.9
ENSMUST00000109647.3 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr4_+_145397238 | 0.11 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr4_+_147056433 | 0.11 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989 |
| chr2_-_111779785 | 0.10 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr16_-_69660606 | 0.10 |
ENSMUST00000076500.14
|
Speer2
|
spermatogenesis associated glutamate (E)-rich protein 2 |
| chr2_-_111820618 | 0.10 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr16_-_19341016 | 0.09 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr7_+_134870237 | 0.09 |
ENSMUST00000210697.2
ENSMUST00000097983.5 |
Nps
|
neuropeptide S |
| chr6_+_24733239 | 0.07 |
ENSMUST00000031690.6
|
Hyal6
|
hyaluronoglucosaminidase 6 |
| chr8_+_40807344 | 0.07 |
ENSMUST00000136835.2
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr2_+_151336095 | 0.07 |
ENSMUST00000089140.13
|
Nsfl1c
|
NSFL1 (p97) cofactor (p47) |
| chr2_-_86257093 | 0.07 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr11_-_116572116 | 0.07 |
ENSMUST00000144398.2
|
St6galnac2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr2_+_37101586 | 0.05 |
ENSMUST00000214897.2
|
Olfr366
|
olfactory receptor 366 |
| chrY_-_35085749 | 0.05 |
ENSMUST00000180170.2
|
Gm20855
|
predicted gene, 20855 |
| chr5_-_23889607 | 0.05 |
ENSMUST00000197985.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr7_-_11414074 | 0.05 |
ENSMUST00000227010.2
ENSMUST00000209638.3 |
Vmn1r72
|
vomeronasal 1 receptor 72 |
| chr2_-_88581690 | 0.04 |
ENSMUST00000215179.3
ENSMUST00000215529.3 |
Olfr1198
|
olfactory receptor 1198 |
| chr14_+_69585036 | 0.04 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr2_-_89779008 | 0.03 |
ENSMUST00000214846.2
|
Olfr1259
|
olfactory receptor 1259 |
| chr14_+_47535717 | 0.03 |
ENSMUST00000166743.9
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr7_+_26456567 | 0.03 |
ENSMUST00000077855.8
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr1_-_126758520 | 0.03 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr4_+_115099237 | 0.03 |
ENSMUST00000118278.2
|
Cyp4a29
|
cytochrome P450, family 4, subfamily a, polypeptide 29 |
| chr11_+_49045204 | 0.02 |
ENSMUST00000216273.2
|
Olfr1394
|
olfactory receptor 1394 |
| chr1_-_186947618 | 0.02 |
ENSMUST00000110945.4
ENSMUST00000183931.8 ENSMUST00000027908.13 |
Spata17
|
spermatogenesis associated 17 |
| chr5_-_103777145 | 0.01 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr15_+_57775595 | 0.01 |
ENSMUST00000022992.13
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr19_-_13828056 | 0.01 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr12_+_31440842 | 0.01 |
ENSMUST00000167432.8
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr15_-_101422054 | 0.01 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr7_-_84661476 | 0.00 |
ENSMUST00000124773.3
ENSMUST00000233725.2 ENSMUST00000233739.2 ENSMUST00000232837.2 |
Vmn2r66
|
vomeronasal 2, receptor 66 |
| chr16_-_58898368 | 0.00 |
ENSMUST00000216495.3
|
Olfr190
|
olfactory receptor 190 |
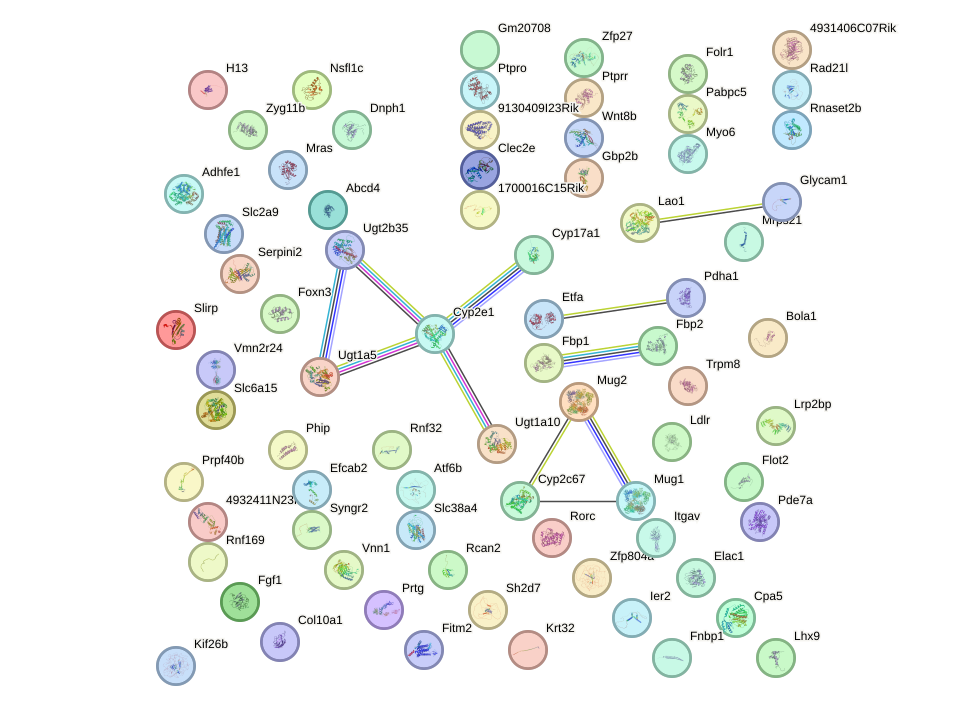
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.5 | 5.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 1.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.4 | 1.7 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.4 | 8.5 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.3 | 0.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.3 | 1.2 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 4.7 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.2 | 0.7 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.2 | 0.9 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 2.4 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.2 | 3.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 6.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 0.8 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.6 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.7 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.5 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 1.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.3 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.8 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.6 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.2 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 1.7 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 1.1 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 1.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 1.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 2.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 3.1 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.0 | 0.4 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.4 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 4.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.8 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 1.3 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.7 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.2 | 0.7 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 0.6 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 1.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 4.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 4.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 6.1 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.1 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.0 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0019002 | GMP binding(GO:0019002) |
| 0.5 | 2.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.5 | 3.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.4 | 2.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.4 | 8.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.4 | 2.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 3.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 5.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 8.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 6.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 1.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.7 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.5 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.6 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 10.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.3 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 4.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.9 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 4.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.6 | 8.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 3.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 5.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 4.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.1 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 2.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |