Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxd9
Z-value: 0.86
Transcription factors associated with Hoxd9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd9
|
ENSMUSG00000043342.10 | homeobox D9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd9 | mm39_v1_chr2_+_74528071_74528097 | 0.28 | 9.9e-02 | Click! |
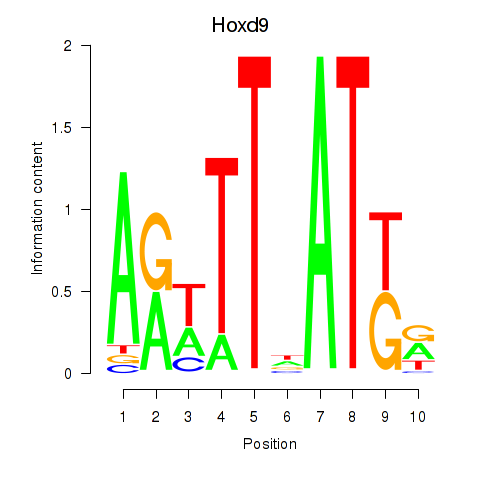
Activity profile of Hoxd9 motif
Sorted Z-values of Hoxd9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_41291634 | 5.89 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr14_+_66043281 | 4.02 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr15_+_6673167 | 3.91 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr3_-_115923098 | 3.86 |
ENSMUST00000196449.5
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr3_-_14843512 | 2.99 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chrX_-_142610371 | 2.81 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chr15_-_98832403 | 2.78 |
ENSMUST00000077577.8
|
Tuba1b
|
tubulin, alpha 1B |
| chr6_-_142418801 | 2.58 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr9_-_64080161 | 2.07 |
ENSMUST00000176299.8
ENSMUST00000130127.8 ENSMUST00000176794.8 ENSMUST00000177045.8 |
Zwilch
|
zwilch kinetochore protein |
| chr3_+_60503051 | 1.82 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chrX_-_165992311 | 1.81 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr16_-_75706161 | 1.77 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr10_-_35587888 | 1.75 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr7_+_89814713 | 1.72 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chrX_-_165992145 | 1.67 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr5_+_117378510 | 1.64 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr12_+_117807224 | 1.64 |
ENSMUST00000021592.16
|
Cdca7l
|
cell division cycle associated 7 like |
| chrX_-_48886577 | 1.63 |
ENSMUST00000033442.14
ENSMUST00000114891.2 |
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr1_-_23948764 | 1.55 |
ENSMUST00000129254.8
|
Smap1
|
small ArfGAP 1 |
| chr2_-_84255602 | 1.39 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr6_+_38918327 | 1.37 |
ENSMUST00000160963.2
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr14_+_66378382 | 1.35 |
ENSMUST00000022620.11
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr2_-_151586063 | 1.32 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr17_-_78991691 | 1.28 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr1_-_138103021 | 1.23 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chrX_-_133652140 | 1.22 |
ENSMUST00000052431.12
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr19_-_56536646 | 1.22 |
ENSMUST00000182276.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr2_+_11176891 | 1.19 |
ENSMUST00000028118.9
|
Prkcq
|
protein kinase C, theta |
| chr9_-_39515420 | 1.17 |
ENSMUST00000042485.11
ENSMUST00000141370.8 |
AW551984
|
expressed sequence AW551984 |
| chr1_+_88139678 | 1.14 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr10_+_89580849 | 1.12 |
ENSMUST00000020112.7
|
Uhrf1bp1l
|
UHRF1 (ICBP90) binding protein 1-like |
| chr6_-_87510200 | 1.09 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr3_-_129834788 | 1.09 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr6_-_69800923 | 1.09 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr16_+_29884153 | 1.06 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr1_-_97689263 | 1.06 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr14_+_43951187 | 1.04 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chrX_-_133652080 | 1.04 |
ENSMUST00000113194.8
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr4_-_129472328 | 1.03 |
ENSMUST00000052835.9
|
Fam167b
|
family with sequence similarity 167, member B |
| chr19_+_56536685 | 1.03 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr5_-_18054702 | 1.01 |
ENSMUST00000165232.8
|
Cd36
|
CD36 molecule |
| chr10_-_93727003 | 0.98 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr1_-_154975376 | 0.96 |
ENSMUST00000055322.6
|
Ier5
|
immediate early response 5 |
| chr12_-_41536430 | 0.91 |
ENSMUST00000043884.6
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr7_+_120450406 | 0.89 |
ENSMUST00000143322.9
ENSMUST00000106488.3 |
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr19_+_45433899 | 0.87 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr7_+_127661807 | 0.87 |
ENSMUST00000064821.14
|
Itgam
|
integrin alpha M |
| chr15_-_37459570 | 0.86 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chrX_-_10083157 | 0.86 |
ENSMUST00000072393.9
ENSMUST00000044598.13 ENSMUST00000073392.11 ENSMUST00000115533.8 ENSMUST00000115532.2 |
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr1_-_138102972 | 0.85 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr6_-_68840015 | 0.84 |
ENSMUST00000103336.2
|
Igkv1-88
|
immunoglobulin kappa chain variable 1-88 |
| chrX_-_133442596 | 0.81 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr11_+_23234644 | 0.80 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chrX_+_55493325 | 0.78 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr6_+_57679621 | 0.75 |
ENSMUST00000050077.15
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr19_-_56536443 | 0.74 |
ENSMUST00000182059.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr12_-_113589576 | 0.72 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr1_-_160079007 | 0.72 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr5_+_145282064 | 0.71 |
ENSMUST00000079268.9
|
Cyp3a57
|
cytochrome P450, family 3, subfamily a, polypeptide 57 |
| chr4_+_5724305 | 0.71 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr12_-_118265103 | 0.69 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr6_+_70192384 | 0.68 |
ENSMUST00000103383.3
|
Igkv6-25
|
immunoglobulin kappa chain variable 6-25 |
| chr3_+_5815863 | 0.65 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr18_+_42669322 | 0.63 |
ENSMUST00000236418.2
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr6_+_83985684 | 0.61 |
ENSMUST00000203803.3
ENSMUST00000204591.3 ENSMUST00000113823.8 ENSMUST00000153860.4 |
Dysf
|
dysferlin |
| chr1_-_128030148 | 0.58 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr11_+_67061908 | 0.57 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr1_+_180158035 | 0.56 |
ENSMUST00000070181.7
|
Itpkb
|
inositol 1,4,5-trisphosphate 3-kinase B |
| chr10_+_74872898 | 0.56 |
ENSMUST00000147802.9
ENSMUST00000020391.13 ENSMUST00000234625.2 |
Rab36
|
RAB36, member RAS oncogene family |
| chr6_+_48514518 | 0.56 |
ENSMUST00000040361.8
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2 |
| chr18_+_31922173 | 0.53 |
ENSMUST00000025106.5
ENSMUST00000234146.2 |
Polr2d
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr10_-_129107354 | 0.50 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr2_+_91357100 | 0.49 |
ENSMUST00000111338.10
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr3_+_32490525 | 0.49 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr5_-_62923463 | 0.46 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_100521343 | 0.45 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr3_+_103078971 | 0.45 |
ENSMUST00000005830.15
|
Bcas2
|
breast carcinoma amplified sequence 2 |
| chr8_-_41469332 | 0.45 |
ENSMUST00000131965.2
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr14_-_75991903 | 0.43 |
ENSMUST00000049168.9
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr16_-_22847760 | 0.41 |
ENSMUST00000039338.13
|
Kng2
|
kininogen 2 |
| chr8_+_85583611 | 0.39 |
ENSMUST00000003906.13
ENSMUST00000109754.2 |
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr11_+_31823096 | 0.38 |
ENSMUST00000155278.2
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr7_+_100435548 | 0.37 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A |
| chr16_-_22847829 | 0.37 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr1_-_171854818 | 0.35 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr5_-_104125192 | 0.34 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr11_+_67061837 | 0.33 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr19_-_57185928 | 0.33 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr6_+_108771840 | 0.33 |
ENSMUST00000204483.2
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr4_-_43710231 | 0.32 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr2_+_83642910 | 0.31 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr19_-_57185988 | 0.30 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr14_-_123151162 | 0.30 |
ENSMUST00000160401.8
|
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr2_-_85966272 | 0.30 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr6_-_98319684 | 0.28 |
ENSMUST00000164491.3
|
Mdfic2
|
MyoD family inhibitor domain containing 2 |
| chr16_-_22847808 | 0.28 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr10_-_18890281 | 0.28 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr7_-_84328553 | 0.26 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr17_+_75742881 | 0.26 |
ENSMUST00000164192.9
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr16_-_65359406 | 0.25 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr3_+_32490300 | 0.25 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr3_+_113824181 | 0.25 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr5_-_69748126 | 0.25 |
ENSMUST00000166298.8
|
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr8_+_66838927 | 0.25 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr1_-_140111138 | 0.24 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chrX_+_41241049 | 0.24 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr3_-_49711706 | 0.23 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr2_-_45000389 | 0.23 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_37537224 | 0.23 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr16_+_21644692 | 0.21 |
ENSMUST00000232240.2
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr3_+_30656204 | 0.21 |
ENSMUST00000192715.6
|
Mynn
|
myoneurin |
| chr7_+_45271229 | 0.20 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr6_+_18848600 | 0.20 |
ENSMUST00000201141.3
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr13_+_49697919 | 0.19 |
ENSMUST00000177948.2
ENSMUST00000021820.14 |
Aspn
|
asporin |
| chr3_+_67799510 | 0.18 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr14_-_56137697 | 0.17 |
ENSMUST00000111325.5
|
Sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr1_-_140111018 | 0.17 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr2_-_152422220 | 0.16 |
ENSMUST00000053180.4
|
Defb19
|
defensin beta 19 |
| chr3_+_35808269 | 0.16 |
ENSMUST00000029257.15
|
Atp11b
|
ATPase, class VI, type 11B |
| chrX_+_113384008 | 0.15 |
ENSMUST00000113371.8
ENSMUST00000040504.12 |
Klhl4
|
kelch-like 4 |
| chr3_-_49711765 | 0.14 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr2_-_76700830 | 0.14 |
ENSMUST00000138542.2
|
Ttn
|
titin |
| chr1_-_82746169 | 0.14 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr3_-_144525255 | 0.14 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr6_+_149226891 | 0.13 |
ENSMUST00000189837.2
|
Resf1
|
retroelement silencing factor 1 |
| chr19_+_34078333 | 0.13 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr5_-_69699932 | 0.12 |
ENSMUST00000202423.2
|
Yipf7
|
Yip1 domain family, member 7 |
| chr5_-_21087023 | 0.12 |
ENSMUST00000118174.8
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr9_-_60594742 | 0.10 |
ENSMUST00000114032.8
ENSMUST00000166168.8 ENSMUST00000132366.2 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr4_+_109092610 | 0.10 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr19_+_39980868 | 0.09 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr3_-_75177378 | 0.09 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr9_-_18297277 | 0.09 |
ENSMUST00000166825.8
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr6_+_83985495 | 0.08 |
ENSMUST00000113821.8
|
Dysf
|
dysferlin |
| chr13_+_93441447 | 0.08 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr10_+_40559249 | 0.08 |
ENSMUST00000058747.4
|
Mettl24
|
methyltransferase like 24 |
| chr19_-_57185861 | 0.08 |
ENSMUST00000111550.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr2_-_88534814 | 0.07 |
ENSMUST00000216928.2
ENSMUST00000216977.2 |
Olfr1196
|
olfactory receptor 1196 |
| chr5_+_136982100 | 0.07 |
ENSMUST00000111094.8
ENSMUST00000111097.8 |
Fis1
|
fission, mitochondrial 1 |
| chr2_-_115895202 | 0.06 |
ENSMUST00000110906.9
|
Meis2
|
Meis homeobox 2 |
| chr10_+_129153986 | 0.06 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr2_+_85648823 | 0.06 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr7_+_107679062 | 0.05 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
| chr6_+_41095752 | 0.05 |
ENSMUST00000103269.3
|
Trbv12-2
|
T cell receptor beta, variable 12-2 |
| chr4_+_109092459 | 0.04 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chrM_+_11735 | 0.04 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr2_-_87467879 | 0.03 |
ENSMUST00000216082.2
|
Olfr1132
|
olfactory receptor 1132 |
| chr10_-_81102740 | 0.02 |
ENSMUST00000046114.5
|
Mrpl54
|
mitochondrial ribosomal protein L54 |
| chr13_-_100453124 | 0.02 |
ENSMUST00000042220.3
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr3_+_138058139 | 0.02 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr9_+_43222104 | 0.02 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr18_+_32200781 | 0.01 |
ENSMUST00000025243.5
ENSMUST00000212675.2 |
Iws1
|
IWS1, SUPT6 interacting protein |
| chr17_+_78815493 | 0.01 |
ENSMUST00000024880.11
ENSMUST00000232859.2 |
Vit
|
vitrin |
| chr8_-_62576140 | 0.00 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr2_+_88470886 | 0.00 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr10_+_23952398 | 0.00 |
ENSMUST00000051133.6
|
Taar8a
|
trace amine-associated receptor 8A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.5 | 2.1 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.4 | 1.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.4 | 1.1 | GO:2000978 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.3 | 1.4 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.3 | 3.9 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.3 | 1.7 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.3 | 1.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.7 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 2.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 1.0 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 3.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 0.7 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.2 | 0.5 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.1 | 1.2 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 0.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.9 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 1.0 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 0.4 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.6 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 0.3 | GO:0034148 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 1.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 5.7 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 1.2 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 2.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 1.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 1.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 3.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 1.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.8 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 3.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 1.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 5.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 2.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 1.3 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 2.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 1.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 2.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 3.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0005662 | Prp19 complex(GO:0000974) DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.7 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.5 | 1.4 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.4 | 3.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 1.1 | GO:0071820 | N-box binding(GO:0071820) |
| 0.3 | 3.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 0.9 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.3 | 1.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 2.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 1.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 1.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.7 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 1.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 2.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 2.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 3.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 1.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 1.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.6 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 1.4 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 2.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 7.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 3.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.9 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 4.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 2.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |