Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
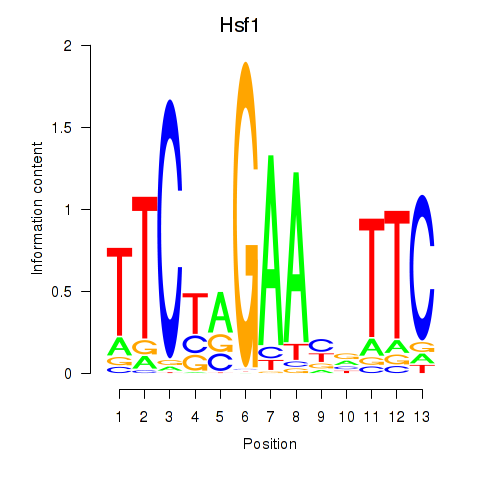
Results for Hsf1
Z-value: 0.63
Transcription factors associated with Hsf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf1
|
ENSMUSG00000022556.12 | heat shock factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf1 | mm39_v1_chr15_+_76361604_76361774 | 0.46 | 4.3e-03 | Click! |
Activity profile of Hsf1 motif
Sorted Z-values of Hsf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_110662256 | 3.50 |
ENSMUST00000149189.2
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr11_+_87684299 | 2.92 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr5_-_149559636 | 2.73 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr12_-_110662677 | 2.73 |
ENSMUST00000124156.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr12_-_110662723 | 2.70 |
ENSMUST00000021698.13
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr12_-_110662765 | 2.61 |
ENSMUST00000094361.11
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr5_-_149559667 | 2.47 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr17_+_48606948 | 2.25 |
ENSMUST00000233092.2
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr3_+_79498663 | 2.15 |
ENSMUST00000029382.13
|
Ppid
|
peptidylprolyl isomerase D (cyclophilin D) |
| chr17_+_48607405 | 2.14 |
ENSMUST00000170941.3
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr6_-_51446752 | 1.80 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr7_-_110462446 | 1.76 |
ENSMUST00000033050.5
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr5_-_149559792 | 1.76 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr5_-_129864202 | 1.63 |
ENSMUST00000136507.4
|
Psph
|
phosphoserine phosphatase |
| chr5_-_36853281 | 1.51 |
ENSMUST00000031091.13
ENSMUST00000140653.2 |
D5Ertd579e
|
DNA segment, Chr 5, ERATO Doi 579, expressed |
| chrX_+_41238193 | 1.50 |
ENSMUST00000115073.9
ENSMUST00000115072.8 |
Stag2
|
stromal antigen 2 |
| chr9_+_119939414 | 1.48 |
ENSMUST00000035106.12
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr5_+_129864044 | 1.43 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr12_-_104964936 | 1.41 |
ENSMUST00000109927.2
ENSMUST00000095439.11 |
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr6_-_51446850 | 1.33 |
ENSMUST00000069949.13
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr1_-_160040286 | 1.32 |
ENSMUST00000195654.2
ENSMUST00000014370.11 |
Cacybp
|
calcyclin binding protein |
| chr9_+_18203640 | 1.32 |
ENSMUST00000217031.2
|
Chordc1
|
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| chr2_+_157401998 | 1.27 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr5_+_29940686 | 1.26 |
ENSMUST00000008733.15
|
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr6_+_51447317 | 1.22 |
ENSMUST00000094623.10
|
Cbx3
|
chromobox 3 |
| chr9_+_123902143 | 1.21 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr9_+_18203421 | 1.21 |
ENSMUST00000001825.9
|
Chordc1
|
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| chr14_-_66071412 | 1.13 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr8_+_88999031 | 1.10 |
ENSMUST00000169037.9
|
Adcy7
|
adenylate cyclase 7 |
| chr9_+_18203558 | 1.04 |
ENSMUST00000213605.2
|
Chordc1
|
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| chr17_+_47904355 | 1.04 |
ENSMUST00000182209.8
|
Ccnd3
|
cyclin D3 |
| chr6_-_136918885 | 1.01 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr11_+_22940519 | 0.96 |
ENSMUST00000173867.8
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chr17_+_47904441 | 0.95 |
ENSMUST00000182874.3
|
Ccnd3
|
cyclin D3 |
| chr17_-_45883421 | 0.95 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr1_-_55127312 | 0.94 |
ENSMUST00000127861.8
ENSMUST00000144077.3 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr5_+_30486375 | 0.92 |
ENSMUST00000101448.5
|
Drc1
|
dynein regulatory complex subunit 1 |
| chrX_-_60229164 | 0.88 |
ENSMUST00000166381.3
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr12_+_105750952 | 0.87 |
ENSMUST00000109901.9
ENSMUST00000168186.8 ENSMUST00000163473.8 ENSMUST00000170540.8 ENSMUST00000166735.8 ENSMUST00000170002.8 |
Papola
|
poly (A) polymerase alpha |
| chr16_-_87292592 | 0.87 |
ENSMUST00000176750.2
ENSMUST00000175977.8 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr9_-_103357564 | 0.84 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr11_+_22940599 | 0.83 |
ENSMUST00000020562.5
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chr16_-_22084700 | 0.83 |
ENSMUST00000161286.8
|
Tra2b
|
transformer 2 beta |
| chr5_+_29940935 | 0.80 |
ENSMUST00000114839.8
ENSMUST00000198694.5 ENSMUST00000012734.10 ENSMUST00000196528.5 |
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr11_-_5049036 | 0.79 |
ENSMUST00000102930.10
ENSMUST00000093365.12 ENSMUST00000073308.11 |
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr11_+_83328503 | 0.78 |
ENSMUST00000037378.6
|
1700020L24Rik
|
RIKEN cDNA 1700020L24 gene |
| chr5_-_123887434 | 0.77 |
ENSMUST00000182955.8
ENSMUST00000182489.8 ENSMUST00000183147.9 ENSMUST00000050827.14 ENSMUST00000057795.12 ENSMUST00000111515.8 ENSMUST00000182309.8 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr8_-_70411054 | 0.74 |
ENSMUST00000211960.2
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr16_-_87292711 | 0.72 |
ENSMUST00000176041.8
ENSMUST00000026704.14 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr11_-_23447866 | 0.69 |
ENSMUST00000128559.2
ENSMUST00000147157.8 ENSMUST00000109539.8 |
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr2_+_119727689 | 0.68 |
ENSMUST00000046717.13
ENSMUST00000079934.12 ENSMUST00000110774.8 ENSMUST00000110773.9 ENSMUST00000156510.2 |
Mga
|
MAX gene associated |
| chr10_-_86541349 | 0.67 |
ENSMUST00000020238.14
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chr7_+_92210348 | 0.64 |
ENSMUST00000032842.13
ENSMUST00000085017.5 |
Ccdc90b
|
coiled-coil domain containing 90B |
| chr5_-_149559737 | 0.63 |
ENSMUST00000200805.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_-_55127183 | 0.62 |
ENSMUST00000027123.15
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr15_-_31601652 | 0.61 |
ENSMUST00000161266.2
|
Cct5
|
chaperonin containing Tcp1, subunit 5 (epsilon) |
| chr8_+_84335176 | 0.53 |
ENSMUST00000212300.2
|
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr9_-_110818679 | 0.53 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chrX_-_139857424 | 0.52 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr6_+_29853745 | 0.52 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr11_-_23448030 | 0.49 |
ENSMUST00000020529.13
|
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr11_-_5049223 | 0.48 |
ENSMUST00000079949.13
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr7_+_110628158 | 0.48 |
ENSMUST00000005749.6
|
Ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr1_+_91178288 | 0.48 |
ENSMUST00000171112.8
ENSMUST00000191533.2 |
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr19_-_7017295 | 0.48 |
ENSMUST00000025918.9
|
Stip1
|
stress-induced phosphoprotein 1 |
| chr6_-_128415640 | 0.47 |
ENSMUST00000032508.11
|
Fkbp4
|
FK506 binding protein 4 |
| chr1_+_179788675 | 0.46 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr15_-_81284244 | 0.42 |
ENSMUST00000172107.8
ENSMUST00000169204.2 ENSMUST00000163382.2 |
St13
|
suppression of tumorigenicity 13 |
| chr10_+_59159118 | 0.42 |
ENSMUST00000009789.15
ENSMUST00000092512.11 ENSMUST00000105466.3 |
P4ha1
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
| chr15_-_81283795 | 0.40 |
ENSMUST00000023039.15
|
St13
|
suppression of tumorigenicity 13 |
| chr4_+_13784749 | 0.38 |
ENSMUST00000098256.4
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr9_-_105372235 | 0.37 |
ENSMUST00000176190.8
ENSMUST00000163879.9 ENSMUST00000112558.10 ENSMUST00000176363.9 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr19_-_29790352 | 0.37 |
ENSMUST00000099525.5
|
Ranbp6
|
RAN binding protein 6 |
| chr11_+_6510167 | 0.36 |
ENSMUST00000109722.9
|
Ccm2
|
cerebral cavernous malformation 2 |
| chr2_+_163535925 | 0.34 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr7_+_35285657 | 0.34 |
ENSMUST00000040844.16
ENSMUST00000188906.7 ENSMUST00000186245.7 ENSMUST00000190503.7 |
Ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr16_-_20245138 | 0.33 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr15_-_100301124 | 0.33 |
ENSMUST00000124324.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr7_-_89590230 | 0.32 |
ENSMUST00000075010.12
|
Hikeshi
|
heat shock protein nuclear import factor |
| chr3_+_117368483 | 0.31 |
ENSMUST00000039564.11
ENSMUST00000238937.2 |
Plppr5
|
phospholipid phosphatase related 5 |
| chr12_+_117807607 | 0.30 |
ENSMUST00000176735.8
ENSMUST00000177339.2 |
Cdca7l
|
cell division cycle associated 7 like |
| chrX_-_149440388 | 0.29 |
ENSMUST00000151403.9
ENSMUST00000087253.11 ENSMUST00000112709.8 ENSMUST00000163969.8 ENSMUST00000087258.10 |
Tro
|
trophinin |
| chr16_-_88548523 | 0.29 |
ENSMUST00000053149.4
|
Krtap13
|
keratin associated protein 13 |
| chr1_+_57813759 | 0.27 |
ENSMUST00000167971.8
ENSMUST00000170139.8 ENSMUST00000171699.8 ENSMUST00000164302.8 |
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr6_-_72416531 | 0.27 |
ENSMUST00000205335.2
ENSMUST00000206692.2 ENSMUST00000059472.10 |
Mat2a
|
methionine adenosyltransferase II, alpha |
| chr9_+_22322802 | 0.26 |
ENSMUST00000058868.9
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr5_-_143831842 | 0.25 |
ENSMUST00000079624.12
ENSMUST00000110717.9 |
Ankrd61
|
ankyrin repeat domain 61 |
| chr12_+_24881582 | 0.24 |
ENSMUST00000221952.2
ENSMUST00000078902.8 ENSMUST00000110942.11 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr13_+_44883270 | 0.24 |
ENSMUST00000172830.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr13_+_44882998 | 0.23 |
ENSMUST00000174068.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr16_-_20245071 | 0.23 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr12_-_104718159 | 0.22 |
ENSMUST00000041987.7
|
Dicer1
|
dicer 1, ribonuclease type III |
| chr15_+_81284333 | 0.21 |
ENSMUST00000163754.9
ENSMUST00000041609.11 |
Xpnpep3
|
X-prolyl aminopeptidase 3, mitochondrial |
| chr7_-_89590334 | 0.20 |
ENSMUST00000207309.2
ENSMUST00000130609.3 |
Hikeshi
|
heat shock protein nuclear import factor |
| chr16_-_88571089 | 0.19 |
ENSMUST00000054223.4
|
2310057N15Rik
|
RIKEN cDNA 2310057N15 gene |
| chr16_+_88555732 | 0.19 |
ENSMUST00000089111.5
|
2310034C09Rik
|
RIKEN cDNA 2310034C09 gene |
| chr17_-_57181420 | 0.19 |
ENSMUST00000043062.5
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr11_-_69768875 | 0.19 |
ENSMUST00000178597.3
|
Tmem95
|
transmembrane protein 95 |
| chr5_-_66309244 | 0.16 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr11_-_116226175 | 0.16 |
ENSMUST00000036215.8
|
Foxj1
|
forkhead box J1 |
| chr5_-_86616849 | 0.15 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr7_+_140425460 | 0.15 |
ENSMUST00000035300.7
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr6_-_128803182 | 0.14 |
ENSMUST00000204756.3
ENSMUST00000204394.3 ENSMUST00000204423.3 ENSMUST00000204677.2 ENSMUST00000205130.3 ENSMUST00000174544.2 ENSMUST00000172887.8 ENSMUST00000032472.11 |
Gm44511
Klrb1b
|
predicted gene 44511 killer cell lectin-like receptor subfamily B member 1B |
| chr5_+_66833434 | 0.14 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr11_-_5049082 | 0.09 |
ENSMUST00000063232.7
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chrX_-_149440362 | 0.09 |
ENSMUST00000148604.2
|
Tro
|
trophinin |
| chr18_-_36899245 | 0.08 |
ENSMUST00000061522.8
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr7_-_89590404 | 0.08 |
ENSMUST00000153470.9
|
Hikeshi
|
heat shock protein nuclear import factor |
| chr17_+_34636321 | 0.08 |
ENSMUST00000142317.8
|
BC051142
|
cDNA sequence BC051142 |
| chr14_-_66071337 | 0.07 |
ENSMUST00000225853.2
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr7_-_35285001 | 0.07 |
ENSMUST00000069912.6
|
Rgs9bp
|
regulator of G-protein signalling 9 binding protein |
| chr19_+_13595285 | 0.06 |
ENSMUST00000216688.3
|
Olfr1487
|
olfactory receptor 1487 |
| chr17_+_28749808 | 0.06 |
ENSMUST00000233837.2
ENSMUST00000025060.4 |
Armc12
|
armadillo repeat containing 12 |
| chr17_+_25381414 | 0.05 |
ENSMUST00000073277.12
ENSMUST00000182621.8 |
Ccdc154
|
coiled-coil domain containing 154 |
| chr15_+_88484484 | 0.04 |
ENSMUST00000066949.9
|
Zdhhc25
|
zinc finger, DHHC domain containing 25 |
| chr7_-_99002430 | 0.04 |
ENSMUST00000094154.6
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
| chr5_+_149121458 | 0.03 |
ENSMUST00000122160.8
ENSMUST00000100410.10 ENSMUST00000119685.8 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr16_-_55755160 | 0.03 |
ENSMUST00000122280.8
ENSMUST00000121703.3 |
Cep97
|
centrosomal protein 97 |
| chr3_+_117368876 | 0.03 |
ENSMUST00000106473.5
|
Plppr5
|
phospholipid phosphatase related 5 |
| chr2_+_72128239 | 0.03 |
ENSMUST00000144111.2
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr16_-_64422716 | 0.03 |
ENSMUST00000209382.3
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr16_-_94657531 | 0.02 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr11_+_104576528 | 0.02 |
ENSMUST00000148007.3
ENSMUST00000212287.2 |
Gm11639
|
predicted gene 11639 |
| chr12_+_87862140 | 0.01 |
ENSMUST00000220585.2
|
Eif1ad4
|
eukaryotic translation initiation factor 1A domain containing 4 |
| chr12_-_72283465 | 0.01 |
ENSMUST00000021497.16
ENSMUST00000137990.2 |
Rtn1
|
reticulon 1 |
| chrX_+_70707271 | 0.00 |
ENSMUST00000070449.6
|
Gpr50
|
G-protein-coupled receptor 50 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.5 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 1.2 | 3.6 | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000299) |
| 1.0 | 3.1 | GO:1990428 | miRNA transport(GO:1990428) |
| 1.0 | 2.9 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.8 | 7.6 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.7 | 2.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.5 | 1.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.4 | 1.2 | GO:1902567 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.4 | 1.5 | GO:0036233 | glycine import(GO:0036233) |
| 0.4 | 5.3 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.3 | 0.9 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.3 | 2.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 1.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 1.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.5 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.8 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.3 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 1.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 1.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 1.8 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 2.0 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.3 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.7 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 1.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.2 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.7 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 1.3 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.9 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.9 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.3 | 5.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 3.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 1.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 2.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 1.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.3 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 2.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.5 | GO:0002135 | CTP binding(GO:0002135) |
| 0.7 | 7.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 0.8 | GO:0032564 | dATP binding(GO:0032564) |
| 0.4 | 1.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.3 | 2.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 1.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 3.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 1.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 1.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 0.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 5.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 1.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 3.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 2.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 2.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.8 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 5.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 3.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |