Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
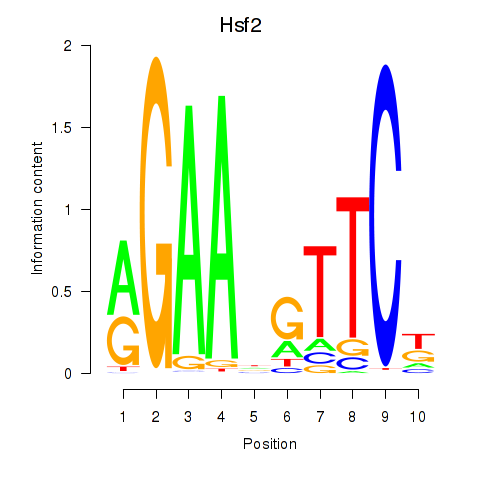
Results for Hsf2
Z-value: 0.76
Transcription factors associated with Hsf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf2
|
ENSMUSG00000019878.9 | heat shock factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf2 | mm39_v1_chr10_+_57362512_57362523 | -0.21 | 2.1e-01 | Click! |
Activity profile of Hsf2 motif
Sorted Z-values of Hsf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_31348576 | 4.15 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr16_+_35977170 | 3.83 |
ENSMUST00000079184.6
|
Stfa2l1
|
stefin A2 like 1 |
| chr9_+_98372575 | 3.65 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr16_-_36228798 | 3.41 |
ENSMUST00000023619.8
|
Stfa2
|
stefin A2 |
| chr16_-_36275739 | 3.18 |
ENSMUST00000068182.3
|
Stfa3
|
stefin A3 |
| chr16_-_36188086 | 3.08 |
ENSMUST00000096089.3
|
Cstdc5
|
cystatin domain containing 5 |
| chr16_+_36097313 | 3.05 |
ENSMUST00000232150.2
|
Stfa1
|
stefin A1 |
| chr16_+_36097505 | 2.87 |
ENSMUST00000042097.11
|
Stfa1
|
stefin A1 |
| chr7_-_120581535 | 2.59 |
ENSMUST00000033169.9
|
Cdr2
|
cerebellar degeneration-related 2 |
| chr7_+_16515265 | 2.42 |
ENSMUST00000108496.9
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr8_+_85628557 | 2.25 |
ENSMUST00000067060.10
ENSMUST00000239392.2 |
Klf1
|
Kruppel-like factor 1 (erythroid) |
| chr6_-_136918885 | 2.22 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr16_+_36041838 | 2.21 |
ENSMUST00000187183.7
ENSMUST00000187742.7 |
Csta2
|
cystatin A family member 2 |
| chr13_-_37233179 | 2.19 |
ENSMUST00000037491.11
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr2_-_30720345 | 2.12 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr14_-_70867588 | 1.90 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr4_-_141553306 | 1.84 |
ENSMUST00000102481.4
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr16_+_17798292 | 1.80 |
ENSMUST00000075371.5
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr17_+_47904355 | 1.75 |
ENSMUST00000182209.8
|
Ccnd3
|
cyclin D3 |
| chr16_-_16687119 | 1.74 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr19_-_10181243 | 1.73 |
ENSMUST00000142241.2
ENSMUST00000116542.9 ENSMUST00000025651.6 ENSMUST00000156291.2 |
Fen1
|
flap structure specific endonuclease 1 |
| chr3_-_106057077 | 1.73 |
ENSMUST00000149836.2
|
Chil3
|
chitinase-like 3 |
| chr9_+_54606832 | 1.68 |
ENSMUST00000070070.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr14_-_47655621 | 1.64 |
ENSMUST00000180299.8
|
Dlgap5
|
DLG associated protein 5 |
| chr6_-_7692855 | 1.64 |
ENSMUST00000115542.8
ENSMUST00000148349.2 |
Asns
|
asparagine synthetase |
| chr10_-_62343516 | 1.62 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chr17_+_47904441 | 1.62 |
ENSMUST00000182874.3
|
Ccnd3
|
cyclin D3 |
| chr6_-_136918495 | 1.61 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr6_-_7693184 | 1.58 |
ENSMUST00000031766.12
|
Asns
|
asparagine synthetase |
| chrX_+_134934116 | 1.58 |
ENSMUST00000057625.3
|
Arxes1
|
adipocyte-related X-chromosome expressed sequence 1 |
| chr7_+_126810780 | 1.57 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr6_-_136918671 | 1.52 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr6_-_125471666 | 1.51 |
ENSMUST00000032492.9
|
Cd9
|
CD9 antigen |
| chr2_+_130119077 | 1.49 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr4_-_140805613 | 1.45 |
ENSMUST00000030760.15
|
Necap2
|
NECAP endocytosis associated 2 |
| chr6_-_7693110 | 1.44 |
ENSMUST00000126303.8
|
Asns
|
asparagine synthetase |
| chr1_-_66974492 | 1.41 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr15_+_78810919 | 1.40 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr9_-_14292453 | 1.38 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1 |
| chr6_-_136918844 | 1.38 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr1_-_75110511 | 1.38 |
ENSMUST00000027405.6
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr18_+_62086122 | 1.38 |
ENSMUST00000051720.6
ENSMUST00000235860.2 |
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr8_-_124621483 | 1.37 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chr9_+_7558449 | 1.35 |
ENSMUST00000018765.4
|
Mmp8
|
matrix metallopeptidase 8 |
| chr18_+_36797113 | 1.32 |
ENSMUST00000036765.8
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr9_-_70328816 | 1.31 |
ENSMUST00000034742.8
|
Ccnb2
|
cyclin B2 |
| chr9_+_54606798 | 1.29 |
ENSMUST00000154690.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr8_+_106024294 | 1.29 |
ENSMUST00000015003.10
|
E2f4
|
E2F transcription factor 4 |
| chr16_+_36030773 | 1.28 |
ENSMUST00000089628.4
|
Csta3
|
cystatin A family member 3 |
| chr7_+_110371811 | 1.25 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr2_+_152689881 | 1.24 |
ENSMUST00000164120.8
ENSMUST00000178997.8 ENSMUST00000109816.8 |
Tpx2
|
TPX2, microtubule-associated |
| chr15_-_102112657 | 1.21 |
ENSMUST00000231030.2
ENSMUST00000230687.2 ENSMUST00000229514.2 ENSMUST00000229345.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr4_+_63478478 | 1.20 |
ENSMUST00000080336.4
|
Tmem268
|
transmembrane protein 268 |
| chr6_-_142453531 | 1.19 |
ENSMUST00000134191.3
ENSMUST00000239397.2 ENSMUST00000239395.2 ENSMUST00000032373.12 |
Ldhb
|
lactate dehydrogenase B |
| chr9_-_45896663 | 1.18 |
ENSMUST00000214179.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr6_+_41498716 | 1.17 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr1_-_66974694 | 1.17 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr1_+_136395673 | 1.15 |
ENSMUST00000189413.7
ENSMUST00000047817.12 |
Kif14
|
kinesin family member 14 |
| chr17_+_48606948 | 1.13 |
ENSMUST00000233092.2
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr2_+_29951859 | 1.09 |
ENSMUST00000102866.10
|
Set
|
SET nuclear oncogene |
| chr17_-_36149142 | 1.09 |
ENSMUST00000001566.10
|
Tubb5
|
tubulin, beta 5 class I |
| chr17_+_12338161 | 1.09 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr12_-_104964936 | 1.08 |
ENSMUST00000109927.2
ENSMUST00000095439.11 |
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr1_-_167112784 | 1.08 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr9_+_8544143 | 1.04 |
ENSMUST00000050433.8
ENSMUST00000217462.2 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr4_+_63478454 | 1.02 |
ENSMUST00000124332.8
ENSMUST00000150360.8 |
Tmem268
|
transmembrane protein 268 |
| chrX_+_162873183 | 1.01 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chr17_+_47816074 | 1.01 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr2_-_181333597 | 0.99 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr2_+_152689913 | 0.99 |
ENSMUST00000028969.9
|
Tpx2
|
TPX2, microtubule-associated |
| chr11_-_106205320 | 0.98 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr3_-_115923098 | 0.97 |
ENSMUST00000196449.5
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr18_+_36780913 | 0.97 |
ENSMUST00000140061.8
|
Maskbp3
|
multiple ankyrin repeats single KH domain binding protein 3 |
| chr17_+_25235039 | 0.96 |
ENSMUST00000142000.9
ENSMUST00000137386.8 |
Ift140
|
intraflagellar transport 140 |
| chr17_-_36149100 | 0.96 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I |
| chr9_+_119939414 | 0.95 |
ENSMUST00000035106.12
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr18_+_60880149 | 0.95 |
ENSMUST00000236652.2
ENSMUST00000235966.2 |
Rps14
|
ribosomal protein S14 |
| chr7_-_24997393 | 0.94 |
ENSMUST00000005583.12
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr7_+_28050077 | 0.94 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr4_-_46566431 | 0.93 |
ENSMUST00000030021.14
ENSMUST00000107757.8 |
Coro2a
|
coronin, actin binding protein 2A |
| chr1_-_172722589 | 0.92 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr1_-_192955407 | 0.91 |
ENSMUST00000009777.4
|
G0s2
|
G0/G1 switch gene 2 |
| chr6_+_145067457 | 0.91 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr12_+_113115632 | 0.90 |
ENSMUST00000006523.12
ENSMUST00000200553.2 |
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr18_+_50186349 | 0.90 |
ENSMUST00000148159.3
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr12_-_110662723 | 0.88 |
ENSMUST00000021698.13
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr5_+_135916764 | 0.87 |
ENSMUST00000005077.7
|
Hspb1
|
heat shock protein 1 |
| chr2_+_84818538 | 0.86 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr8_+_72889607 | 0.86 |
ENSMUST00000238492.2
|
Tpm4
|
tropomyosin 4 |
| chr2_-_172212426 | 0.86 |
ENSMUST00000109139.8
ENSMUST00000028997.8 ENSMUST00000109140.10 |
Aurka
|
aurora kinase A |
| chr2_+_71219561 | 0.86 |
ENSMUST00000028408.3
|
Hat1
|
histone aminotransferase 1 |
| chr5_-_77457895 | 0.85 |
ENSMUST00000047860.9
|
Noa1
|
nitric oxide associated 1 |
| chr18_-_53551127 | 0.85 |
ENSMUST00000025419.9
|
Ppic
|
peptidylprolyl isomerase C |
| chr2_+_29951766 | 0.84 |
ENSMUST00000149578.8
|
Set
|
SET nuclear oncogene |
| chr7_+_110367375 | 0.83 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr12_-_110662765 | 0.83 |
ENSMUST00000094361.11
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr13_+_20978283 | 0.83 |
ENSMUST00000021757.5
ENSMUST00000221982.2 |
Aoah
|
acyloxyacyl hydrolase |
| chr9_-_110886306 | 0.83 |
ENSMUST00000195968.2
ENSMUST00000111888.3 |
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr13_+_51799268 | 0.83 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr7_-_24997291 | 0.82 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr7_-_19410749 | 0.82 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr8_+_84335176 | 0.81 |
ENSMUST00000212300.2
|
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr12_-_69274936 | 0.81 |
ENSMUST00000221411.2
ENSMUST00000021359.7 |
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr12_-_28685849 | 0.79 |
ENSMUST00000221871.2
|
Rps7
|
ribosomal protein S7 |
| chr2_+_30331839 | 0.78 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr14_+_75373766 | 0.78 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr8_-_106553822 | 0.78 |
ENSMUST00000239468.2
ENSMUST00000041400.6 |
Ranbp10
|
RAN binding protein 10 |
| chr9_+_106080307 | 0.78 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr6_-_83513184 | 0.77 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr19_+_4928699 | 0.77 |
ENSMUST00000006632.8
|
Zdhhc24
|
zinc finger, DHHC domain containing 24 |
| chr1_+_160898283 | 0.75 |
ENSMUST00000028035.14
ENSMUST00000111620.10 ENSMUST00000111618.8 |
Cenpl
|
centromere protein L |
| chr14_+_43951187 | 0.75 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr2_-_154734824 | 0.74 |
ENSMUST00000099173.11
|
Eif2s2
|
eukaryotic translation initiation factor 2, subunit 2 (beta) |
| chr17_-_50401305 | 0.74 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr10_-_62258195 | 0.74 |
ENSMUST00000020277.9
|
Hkdc1
|
hexokinase domain containing 1 |
| chr11_-_100305654 | 0.73 |
ENSMUST00000066489.13
|
P3h4
|
prolyl 3-hydroxylase family member 4 (non-enzymatic) |
| chr7_-_15993110 | 0.72 |
ENSMUST00000168818.2
|
C5ar1
|
complement component 5a receptor 1 |
| chr12_+_104067346 | 0.72 |
ENSMUST00000021495.4
|
Serpina5
|
serine (or cysteine) peptidase inhibitor, clade A, member 5 |
| chr9_+_8544228 | 0.72 |
ENSMUST00000214596.2
|
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr11_-_3454766 | 0.72 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr17_+_47816137 | 0.72 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr7_-_46365108 | 0.71 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr12_-_28685913 | 0.70 |
ENSMUST00000074267.5
|
Rps7
|
ribosomal protein S7 |
| chr2_-_154734702 | 0.70 |
ENSMUST00000166171.8
ENSMUST00000161172.3 |
Eif2s2
|
eukaryotic translation initiation factor 2, subunit 2 (beta) |
| chr17_-_25946370 | 0.70 |
ENSMUST00000170070.3
ENSMUST00000048054.14 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr11_-_100012384 | 0.69 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chr5_-_138169476 | 0.69 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr6_+_135339543 | 0.68 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr17_-_6367692 | 0.68 |
ENSMUST00000232499.2
ENSMUST00000169415.3 |
Dynlt1a
|
dynein light chain Tctex-type 1A |
| chrX_-_156275231 | 0.67 |
ENSMUST00000112529.8
|
Sms
|
spermine synthase |
| chr9_-_42035560 | 0.67 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr11_+_115455260 | 0.67 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr17_+_29171386 | 0.67 |
ENSMUST00000118762.9
ENSMUST00000057174.16 ENSMUST00000232874.2 ENSMUST00000232772.2 ENSMUST00000233334.2 ENSMUST00000150858.2 ENSMUST00000233064.2 |
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr14_+_70337540 | 0.66 |
ENSMUST00000022680.9
|
Bin3
|
bridging integrator 3 |
| chr1_+_40554513 | 0.65 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr10_+_60182630 | 0.64 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr9_+_44318926 | 0.64 |
ENSMUST00000216076.2
ENSMUST00000216867.2 |
Rps25
|
ribosomal protein S25 |
| chr5_+_29940686 | 0.64 |
ENSMUST00000008733.15
|
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr4_-_116982804 | 0.62 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr8_+_75836187 | 0.62 |
ENSMUST00000164309.3
ENSMUST00000212426.2 ENSMUST00000212811.2 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr5_-_137869969 | 0.62 |
ENSMUST00000196162.5
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chr19_-_5416339 | 0.62 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr7_-_12743720 | 0.61 |
ENSMUST00000210282.2
ENSMUST00000172240.2 ENSMUST00000051390.9 ENSMUST00000209997.2 |
Zbtb45
|
zinc finger and BTB domain containing 45 |
| chr10_+_7543260 | 0.61 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr12_+_83567240 | 0.61 |
ENSMUST00000021645.9
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr16_-_87292592 | 0.60 |
ENSMUST00000176750.2
ENSMUST00000175977.8 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr5_+_129864044 | 0.60 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr2_-_150881283 | 0.60 |
ENSMUST00000128627.2
ENSMUST00000066640.5 |
Ninl
Nanp
|
ninein-like N-acetylneuraminic acid phosphatase |
| chr5_+_135916847 | 0.59 |
ENSMUST00000111155.2
|
Hspb1
|
heat shock protein 1 |
| chr2_-_34645241 | 0.59 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr3_+_87989278 | 0.59 |
ENSMUST00000071812.11
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr2_-_121101822 | 0.58 |
ENSMUST00000110647.8
ENSMUST00000110648.8 |
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr6_+_29433247 | 0.58 |
ENSMUST00000101617.9
ENSMUST00000065090.8 |
Flnc
|
filamin C, gamma |
| chr19_-_34143437 | 0.57 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr17_+_34823236 | 0.57 |
ENSMUST00000174041.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr5_-_137145030 | 0.56 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr1_-_133352115 | 0.56 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr5_+_121342544 | 0.56 |
ENSMUST00000031617.13
|
Rpl6
|
ribosomal protein L6 |
| chr3_+_103078971 | 0.56 |
ENSMUST00000005830.15
|
Bcas2
|
breast carcinoma amplified sequence 2 |
| chr11_-_114892706 | 0.55 |
ENSMUST00000092464.10
|
Cd300c2
|
CD300C molecule 2 |
| chr6_+_129374441 | 0.55 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr4_-_117232650 | 0.55 |
ENSMUST00000094853.9
|
Rnf220
|
ring finger protein 220 |
| chr2_+_112069813 | 0.55 |
ENSMUST00000028554.4
|
Lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr10_-_14581203 | 0.54 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr18_+_36662276 | 0.54 |
ENSMUST00000237595.2
|
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr5_-_137856280 | 0.53 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr7_+_127845984 | 0.53 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr13_-_49401617 | 0.53 |
ENSMUST00000119721.2
ENSMUST00000058196.13 |
Susd3
|
sushi domain containing 3 |
| chr11_+_100306523 | 0.53 |
ENSMUST00000001595.10
ENSMUST00000107400.3 |
Fkbp10
|
FK506 binding protein 10 |
| chrX_+_21350783 | 0.52 |
ENSMUST00000089188.9
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr9_-_110886576 | 0.52 |
ENSMUST00000199839.5
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr7_-_141023902 | 0.52 |
ENSMUST00000026580.12
|
Pidd1
|
p53 induced death domain protein 1 |
| chr6_-_51446752 | 0.52 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr16_-_87292711 | 0.52 |
ENSMUST00000176041.8
ENSMUST00000026704.14 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr7_+_3341597 | 0.52 |
ENSMUST00000164553.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_+_25380583 | 0.51 |
ENSMUST00000108403.4
|
B9d2
|
B9 protein domain 2 |
| chr12_-_110662677 | 0.51 |
ENSMUST00000124156.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr12_-_113561594 | 0.51 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr5_+_35156389 | 0.50 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr7_-_141023199 | 0.50 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr10_-_116899664 | 0.50 |
ENSMUST00000218719.2
ENSMUST00000219573.2 ENSMUST00000047672.9 |
Cct2
|
chaperonin containing Tcp1, subunit 2 (beta) |
| chrX_+_55825033 | 0.49 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr19_+_7534838 | 0.49 |
ENSMUST00000141887.8
ENSMUST00000136756.2 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chr17_-_26016039 | 0.48 |
ENSMUST00000165838.9
ENSMUST00000002344.7 |
Metrn
|
meteorin, glial cell differentiation regulator |
| chr16_-_15455141 | 0.48 |
ENSMUST00000023353.4
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr4_+_133829898 | 0.48 |
ENSMUST00000070246.9
ENSMUST00000156750.2 |
Ubxn11
|
UBX domain protein 11 |
| chr5_+_96941284 | 0.47 |
ENSMUST00000200379.5
|
Anxa3
|
annexin A3 |
| chr7_-_126651847 | 0.47 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr6_-_116693849 | 0.47 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr3_+_122522592 | 0.47 |
ENSMUST00000066728.10
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr9_-_44024767 | 0.47 |
ENSMUST00000216511.2
ENSMUST00000056328.6 ENSMUST00000185479.2 |
Rnf26
Gm49380
|
ring finger protein 26 predicted gene, 49380 |
| chr1_-_78465479 | 0.46 |
ENSMUST00000190441.2
ENSMUST00000170217.8 ENSMUST00000188247.7 ENSMUST00000068333.14 |
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr15_-_12824691 | 0.46 |
ENSMUST00000228177.2
ENSMUST00000227299.2 ENSMUST00000057256.5 |
6030458C11Rik
|
RIKEN cDNA 6030458C11 gene |
| chrX_+_55824797 | 0.45 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr5_+_44070486 | 0.45 |
ENSMUST00000122204.3
ENSMUST00000200338.2 |
Gm7879
|
predicted pseudogene 7879 |
| chr15_+_12824901 | 0.45 |
ENSMUST00000169061.8
|
Drosha
|
drosha, ribonuclease type III |
| chr2_+_43638814 | 0.45 |
ENSMUST00000112824.8
ENSMUST00000055776.8 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr5_-_137870001 | 0.45 |
ENSMUST00000164886.2
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chr12_-_55126882 | 0.45 |
ENSMUST00000021406.6
|
2700097O09Rik
|
RIKEN cDNA 2700097O09 gene |
| chr6_+_129374260 | 0.45 |
ENSMUST00000032262.14
|
Clec1b
|
C-type lectin domain family 1, member b |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.8 | 2.4 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.7 | 6.7 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.5 | 1.4 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.4 | 1.6 | GO:0033377 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.4 | 3.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 1.1 | GO:0010138 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.3 | 2.4 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.3 | 1.0 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.3 | 1.9 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.3 | 0.9 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) |
| 0.3 | 0.9 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.3 | 5.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 1.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 0.9 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.3 | 0.8 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.3 | 0.8 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.2 | 1.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.2 | 1.0 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 2.0 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 0.7 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 0.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.9 | GO:0045575 | basophil activation(GO:0045575) |
| 0.2 | 0.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 2.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 0.8 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.2 | 5.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.5 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.2 | 0.2 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 2.4 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 1.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 0.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.8 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.4 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 1.4 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 1.6 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.1 | 0.3 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 1.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.5 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.1 | 2.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.4 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.8 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) primary miRNA processing(GO:0031053) |
| 0.1 | 0.3 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.1 | 0.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.7 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 1.5 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.1 | 1.2 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.1 | 0.9 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 2.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.4 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 0.1 | 0.3 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 1.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.6 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.3 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.3 | GO:0061723 | glycophagy(GO:0061723) |
| 0.1 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.8 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.9 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.8 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 0.7 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 1.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.2 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.1 | 0.2 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 0.2 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.1 | 0.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.0 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.1 | 0.4 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 1.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 3.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 1.0 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 1.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.3 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 1.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.8 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.3 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.1 | 1.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 1.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.8 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.4 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 1.7 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.2 | GO:1903659 | transforming growth factor beta activation(GO:0036363) complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.2 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.3 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.1 | 0.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.2 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.0 | 0.1 | GO:1990051 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 0.3 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.4 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.8 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 7.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 1.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.0 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.8 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.3 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.6 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.5 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.3 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 3.4 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.3 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) strand invasion(GO:0042148) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 3.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.8 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 1.4 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.0 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.1 | GO:0034499 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.9 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 2.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 3.2 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 1.5 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.5 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.5 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0002192 | IRES-dependent translational initiation(GO:0002192) IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.7 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 2.3 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.8 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.3 | 1.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.3 | 0.8 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 2.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 1.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 3.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 0.7 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.2 | 1.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 0.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 1.0 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.2 | 1.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 0.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 1.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.1 | 1.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 5.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 6.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 1.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 1.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.2 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.1 | 0.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.3 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 1.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 3.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 3.3 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 2.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.7 | 6.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 2.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 2.6 | GO:0002135 | CTP binding(GO:0002135) |
| 0.4 | 1.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 1.7 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 1.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 2.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 0.7 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.2 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.7 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 2.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 2.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 3.9 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 1.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 0.8 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.2 | 0.5 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 2.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 1.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 1.5 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 0.8 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.4 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.1 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.4 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.2 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 0.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.4 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 1.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 2.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.3 | GO:0004512 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.1 | 0.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 1.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.5 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 1.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.7 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 3.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 4.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.8 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.8 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.3 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) starch binding(GO:2001070) |
| 0.1 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.4 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 1.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.0 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0034012 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 1.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 2.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 4.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 8.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 5.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 3.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 1.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.7 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 1.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 6.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 6.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.9 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 1.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 6.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |