Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
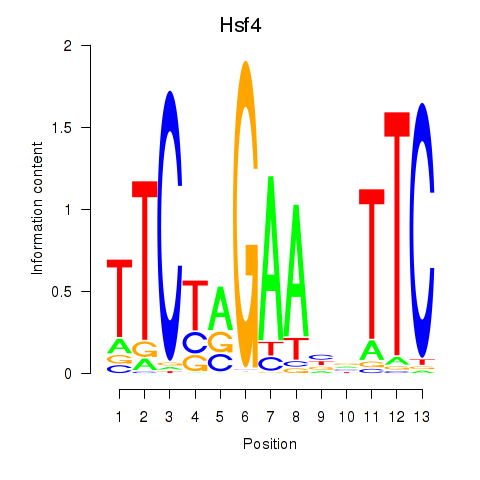
Results for Hsf4
Z-value: 1.39
Transcription factors associated with Hsf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf4
|
ENSMUSG00000033249.11 | heat shock transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf4 | mm39_v1_chr8_+_105996469_105996506 | 0.87 | 1.0e-11 | Click! |
Activity profile of Hsf4 motif
Sorted Z-values of Hsf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_8109346 | 9.09 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr17_-_34218301 | 8.74 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr19_-_8382424 | 8.43 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr7_+_26821266 | 7.04 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr3_-_107850707 | 5.83 |
ENSMUST00000106681.3
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr19_+_58658838 | 5.81 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr8_+_110717062 | 5.79 |
ENSMUST00000001720.14
ENSMUST00000143741.2 |
Tat
|
tyrosine aminotransferase |
| chr3_-_107850666 | 5.61 |
ENSMUST00000106683.8
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr18_-_10706701 | 5.49 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr17_+_35780977 | 5.49 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr7_-_114162125 | 5.44 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr7_-_25239229 | 5.09 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr17_+_35658131 | 5.00 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr17_-_12894716 | 4.99 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr19_-_4087940 | 4.89 |
ENSMUST00000237893.2
ENSMUST00000169613.4 |
Gstp1
|
glutathione S-transferase, pi 1 |
| chr8_-_71990085 | 4.60 |
ENSMUST00000051672.9
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr19_+_4036562 | 4.49 |
ENSMUST00000236224.2
ENSMUST00000236510.2 ENSMUST00000237910.2 ENSMUST00000235612.2 ENSMUST00000054030.8 |
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr5_-_45607463 | 4.33 |
ENSMUST00000197946.5
ENSMUST00000127562.3 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr5_-_45607554 | 4.29 |
ENSMUST00000015950.12
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr5_-_45607485 | 4.29 |
ENSMUST00000154962.8
ENSMUST00000118097.8 ENSMUST00000198258.5 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr15_+_82338247 | 4.21 |
ENSMUST00000230000.2
|
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr5_-_149559667 | 4.01 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr4_+_40722461 | 3.97 |
ENSMUST00000030118.10
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr17_+_35481702 | 3.83 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr5_-_87572060 | 3.66 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr17_-_56428968 | 3.61 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr12_+_112073261 | 3.58 |
ENSMUST00000223412.2
|
Aspg
|
asparaginase |
| chr11_-_106469938 | 3.58 |
ENSMUST00000103070.3
|
Tex2
|
testis expressed gene 2 |
| chrX_-_73067514 | 3.56 |
ENSMUST00000033769.15
ENSMUST00000114352.8 ENSMUST00000068286.12 ENSMUST00000114360.10 ENSMUST00000114354.10 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr17_+_35482063 | 3.50 |
ENSMUST00000172503.3
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chrX_-_37723943 | 3.31 |
ENSMUST00000058265.8
|
C1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr8_-_22888604 | 3.29 |
ENSMUST00000033871.8
|
Slc25a15
|
solute carrier family 25 (mitochondrial carrier ornithine transporter), member 15 |
| chr6_-_128415640 | 3.29 |
ENSMUST00000032508.11
|
Fkbp4
|
FK506 binding protein 4 |
| chrX_-_72759748 | 3.26 |
ENSMUST00000002091.6
|
Bcap31
|
B cell receptor associated protein 31 |
| chr8_-_45835234 | 3.24 |
ENSMUST00000093526.13
|
Fam149a
|
family with sequence similarity 149, member A |
| chr15_+_33083358 | 3.23 |
ENSMUST00000228916.2
ENSMUST00000226483.2 ENSMUST00000228737.2 |
Cpq
|
carboxypeptidase Q |
| chr15_+_33083259 | 3.14 |
ENSMUST00000042167.10
|
Cpq
|
carboxypeptidase Q |
| chr5_-_149559636 | 3.09 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr14_+_66205932 | 2.99 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr16_+_22926504 | 2.91 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr10_-_86541349 | 2.85 |
ENSMUST00000020238.14
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chr8_-_85620537 | 2.76 |
ENSMUST00000003907.14
ENSMUST00000109745.8 ENSMUST00000142748.2 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr5_+_135916764 | 2.73 |
ENSMUST00000005077.7
|
Hspb1
|
heat shock protein 1 |
| chr8_-_95405234 | 2.73 |
ENSMUST00000213043.2
|
Pllp
|
plasma membrane proteolipid |
| chr11_-_23447866 | 2.70 |
ENSMUST00000128559.2
ENSMUST00000147157.8 ENSMUST00000109539.8 |
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr16_+_22926162 | 2.69 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr7_-_119122681 | 2.66 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr5_+_135916847 | 2.56 |
ENSMUST00000111155.2
|
Hspb1
|
heat shock protein 1 |
| chr4_+_155685830 | 2.53 |
ENSMUST00000105608.9
|
Slc35e2
|
solute carrier family 35, member E2 |
| chr11_-_23448030 | 2.47 |
ENSMUST00000020529.13
|
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr15_+_10314173 | 2.44 |
ENSMUST00000127467.3
|
Prlr
|
prolactin receptor |
| chr7_+_121666388 | 2.44 |
ENSMUST00000033158.6
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr17_+_35643818 | 2.39 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chrX_+_21581135 | 2.37 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr5_-_149559792 | 2.35 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr17_+_35643853 | 2.26 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr19_-_7780025 | 2.21 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr13_-_23758370 | 2.19 |
ENSMUST00000105106.2
|
H2bc7
|
H2B clustered histone 7 |
| chr5_-_139799953 | 2.15 |
ENSMUST00000044002.10
|
Tmem184a
|
transmembrane protein 184a |
| chr6_-_59001455 | 2.14 |
ENSMUST00000089860.12
|
Fam13a
|
family with sequence similarity 13, member A |
| chr8_-_71308229 | 2.14 |
ENSMUST00000212086.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr12_-_110662256 | 2.12 |
ENSMUST00000149189.2
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr4_-_107541421 | 2.12 |
ENSMUST00000069271.5
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr16_-_94657531 | 2.10 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr19_-_44057800 | 2.02 |
ENSMUST00000170801.8
|
Erlin1
|
ER lipid raft associated 1 |
| chrX_-_73067351 | 1.99 |
ENSMUST00000114353.10
ENSMUST00000101458.9 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr19_-_7779943 | 1.97 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr10_+_81104006 | 1.93 |
ENSMUST00000057798.9
ENSMUST00000219304.2 |
Apba3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr6_-_83654789 | 1.91 |
ENSMUST00000037882.8
|
Cd207
|
CD207 antigen |
| chr4_+_40722911 | 1.90 |
ENSMUST00000164233.8
ENSMUST00000137246.8 ENSMUST00000125442.8 |
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr16_-_84632383 | 1.89 |
ENSMUST00000114191.8
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr2_-_110144869 | 1.88 |
ENSMUST00000133608.2
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr12_-_110662723 | 1.87 |
ENSMUST00000021698.13
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr12_-_110662765 | 1.85 |
ENSMUST00000094361.11
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr15_-_81283795 | 1.84 |
ENSMUST00000023039.15
|
St13
|
suppression of tumorigenicity 13 |
| chr9_-_105372330 | 1.80 |
ENSMUST00000038118.15
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chr4_+_155646807 | 1.79 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chr17_-_13135232 | 1.78 |
ENSMUST00000079121.4
|
Mrpl18
|
mitochondrial ribosomal protein L18 |
| chr9_-_105372235 | 1.76 |
ENSMUST00000176190.8
ENSMUST00000163879.9 ENSMUST00000112558.10 ENSMUST00000176363.9 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr7_+_136496328 | 1.76 |
ENSMUST00000081510.4
|
Mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chrX_-_60229164 | 1.75 |
ENSMUST00000166381.3
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr7_-_46365108 | 1.73 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr5_+_92702893 | 1.70 |
ENSMUST00000146417.9
|
Fam47e
|
family with sequence similarity 47, member E |
| chr9_-_53617508 | 1.70 |
ENSMUST00000068449.4
|
Rab39
|
RAB39, member RAS oncogene family |
| chr4_+_148225139 | 1.65 |
ENSMUST00000140049.8
ENSMUST00000105707.2 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr11_-_59054107 | 1.64 |
ENSMUST00000069631.3
|
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr15_-_81284244 | 1.63 |
ENSMUST00000172107.8
ENSMUST00000169204.2 ENSMUST00000163382.2 |
St13
|
suppression of tumorigenicity 13 |
| chr19_-_44058129 | 1.62 |
ENSMUST00000169092.2
|
Erlin1
|
ER lipid raft associated 1 |
| chr12_-_110662677 | 1.62 |
ENSMUST00000124156.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr17_-_35265514 | 1.61 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5 |
| chrX_+_138701544 | 1.60 |
ENSMUST00000054889.4
|
Cldn2
|
claudin 2 |
| chr7_+_140425460 | 1.59 |
ENSMUST00000035300.7
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr17_+_31783708 | 1.58 |
ENSMUST00000097352.11
ENSMUST00000237248.2 ENSMUST00000235869.2 ENSMUST00000175806.9 |
Pknox1
|
Pbx/knotted 1 homeobox |
| chr6_+_114620067 | 1.58 |
ENSMUST00000169310.10
ENSMUST00000182169.8 ENSMUST00000183165.8 ENSMUST00000182098.8 ENSMUST00000182793.8 ENSMUST00000182902.8 ENSMUST00000182428.8 ENSMUST00000182035.8 |
Atg7
|
autophagy related 7 |
| chr5_+_92479687 | 1.58 |
ENSMUST00000125462.8
ENSMUST00000113083.9 ENSMUST00000121096.8 |
Art3
|
ADP-ribosyltransferase 3 |
| chr2_+_130247203 | 1.57 |
ENSMUST00000028898.5
|
1700020A23Rik
|
RIKEN cDNA 1700020A23 gene |
| chr16_-_84632439 | 1.57 |
ENSMUST00000138279.2
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr11_-_5492175 | 1.56 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr6_-_124865155 | 1.55 |
ENSMUST00000024044.7
|
Cd4
|
CD4 antigen |
| chr10_+_76398138 | 1.54 |
ENSMUST00000105414.2
|
Spatc1l
|
spermatogenesis and centriole associated 1 like |
| chr7_+_43847615 | 1.54 |
ENSMUST00000085450.4
|
Klk1b3
|
kallikrein 1-related peptidase b3 |
| chr5_-_66309244 | 1.53 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chrX_+_70408351 | 1.52 |
ENSMUST00000146213.8
ENSMUST00000114601.8 ENSMUST00000015358.8 |
Mtmr1
|
myotubularin related protein 1 |
| chr7_+_119495058 | 1.51 |
ENSMUST00000106518.9
ENSMUST00000207270.2 ENSMUST00000208424.2 ENSMUST00000208202.2 ENSMUST00000054440.11 |
Lyrm1
|
LYR motif containing 1 |
| chr7_-_30292351 | 1.51 |
ENSMUST00000108151.3
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr6_-_59001325 | 1.50 |
ENSMUST00000173193.2
|
Fam13a
|
family with sequence similarity 13, member A |
| chr10_+_81104041 | 1.50 |
ENSMUST00000218742.2
|
Apba3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr15_-_4025190 | 1.48 |
ENSMUST00000046633.10
|
AW549877
|
expressed sequence AW549877 |
| chr17_-_45903494 | 1.47 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr5_-_50216249 | 1.47 |
ENSMUST00000030971.7
|
Adgra3
|
adhesion G protein-coupled receptor A3 |
| chr3_+_79498663 | 1.45 |
ENSMUST00000029382.13
|
Ppid
|
peptidylprolyl isomerase D (cyclophilin D) |
| chr5_-_139799780 | 1.45 |
ENSMUST00000146780.3
|
Tmem184a
|
transmembrane protein 184a |
| chr18_-_3337467 | 1.44 |
ENSMUST00000154135.8
ENSMUST00000142690.2 ENSMUST00000025069.11 ENSMUST00000165086.8 ENSMUST00000082141.12 ENSMUST00000149803.8 |
Crem
|
cAMP responsive element modulator |
| chr5_-_123887434 | 1.44 |
ENSMUST00000182955.8
ENSMUST00000182489.8 ENSMUST00000183147.9 ENSMUST00000050827.14 ENSMUST00000057795.12 ENSMUST00000111515.8 ENSMUST00000182309.8 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr7_+_43874752 | 1.44 |
ENSMUST00000075162.5
|
Klk1
|
kallikrein 1 |
| chr2_+_130247159 | 1.43 |
ENSMUST00000188900.2
ENSMUST00000110281.2 ENSMUST00000189961.2 |
Gm28372
1700020A23Rik
|
predicted gene 28372 RIKEN cDNA 1700020A23 gene |
| chr1_+_165312738 | 1.42 |
ENSMUST00000111440.8
ENSMUST00000027852.15 ENSMUST00000111439.8 |
Adcy10
|
adenylate cyclase 10 |
| chr10_+_76398106 | 1.41 |
ENSMUST00000009259.5
|
Spatc1l
|
spermatogenesis and centriole associated 1 like |
| chr8_+_34621717 | 1.41 |
ENSMUST00000239436.2
ENSMUST00000033933.8 |
Saraf
|
store-operated calcium entry-associated regulatory factor |
| chr1_-_166066298 | 1.41 |
ENSMUST00000038782.4
ENSMUST00000194057.6 |
Mael
|
maelstrom spermatogenic transposon silencer |
| chr6_-_33037107 | 1.40 |
ENSMUST00000115091.2
ENSMUST00000127666.8 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr4_+_152262583 | 1.40 |
ENSMUST00000075363.10
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr9_+_119978773 | 1.40 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr6_+_114620054 | 1.40 |
ENSMUST00000032457.17
|
Atg7
|
autophagy related 7 |
| chr3_+_30847024 | 1.39 |
ENSMUST00000029256.9
|
Sec62
|
SEC62 homolog (S. cerevisiae) |
| chr11_-_51891259 | 1.39 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr16_-_84632513 | 1.37 |
ENSMUST00000023608.14
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr4_+_155686311 | 1.35 |
ENSMUST00000118607.2
|
Slc35e2
|
solute carrier family 35, member E2 |
| chr11_+_83599841 | 1.33 |
ENSMUST00000001009.14
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr17_+_53786240 | 1.33 |
ENSMUST00000017975.7
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr7_+_43645253 | 1.31 |
ENSMUST00000007156.5
|
Klk1b11
|
kallikrein 1-related peptidase b11 |
| chr7_+_141988714 | 1.29 |
ENSMUST00000118276.8
ENSMUST00000105976.8 ENSMUST00000097939.9 |
Syt8
|
synaptotagmin VIII |
| chr7_-_119494918 | 1.29 |
ENSMUST00000059851.14
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr11_-_12362136 | 1.29 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr2_-_168072493 | 1.28 |
ENSMUST00000109193.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr6_-_114898739 | 1.28 |
ENSMUST00000032459.14
|
Vgll4
|
vestigial like family member 4 |
| chr2_-_168072295 | 1.28 |
ENSMUST00000154111.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr7_-_23936650 | 1.26 |
ENSMUST00000206960.2
|
Zfp109
|
zinc finger protein 109 |
| chr5_-_149559737 | 1.25 |
ENSMUST00000200805.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr17_+_3376827 | 1.25 |
ENSMUST00000169838.9
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr15_-_82238432 | 1.25 |
ENSMUST00000230494.2
ENSMUST00000023085.7 |
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr5_-_92823111 | 1.23 |
ENSMUST00000151180.8
ENSMUST00000150359.2 |
Ccdc158
|
coiled-coil domain containing 158 |
| chr3_+_85948030 | 1.22 |
ENSMUST00000238545.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr14_+_54014387 | 1.21 |
ENSMUST00000103670.4
|
Trav3-4
|
T cell receptor alpha variable 3-4 |
| chr18_-_3337679 | 1.21 |
ENSMUST00000150235.8
ENSMUST00000154470.8 |
Crem
|
cAMP responsive element modulator |
| chr11_+_73090270 | 1.21 |
ENSMUST00000006105.7
|
Shpk
|
sedoheptulokinase |
| chr16_+_35803674 | 1.19 |
ENSMUST00000004054.13
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr6_+_125026865 | 1.18 |
ENSMUST00000112413.8
|
Acrbp
|
proacrosin binding protein |
| chr7_+_43874854 | 1.18 |
ENSMUST00000206144.2
|
Klk1
|
kallikrein 1 |
| chr4_-_152073448 | 1.16 |
ENSMUST00000036680.8
|
Thap3
|
THAP domain containing, apoptosis associated protein 3 |
| chr1_-_89942299 | 1.16 |
ENSMUST00000086882.8
ENSMUST00000097656.10 |
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr13_+_81805782 | 1.15 |
ENSMUST00000224300.2
ENSMUST00000224592.2 |
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr9_+_18203421 | 1.14 |
ENSMUST00000001825.9
|
Chordc1
|
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| chr9_+_106377153 | 1.13 |
ENSMUST00000164965.3
|
Iqcf1
|
IQ motif containing F1 |
| chr5_+_29940935 | 1.12 |
ENSMUST00000114839.8
ENSMUST00000198694.5 ENSMUST00000012734.10 ENSMUST00000196528.5 |
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr5_+_105879914 | 1.12 |
ENSMUST00000154807.2
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr2_+_109523908 | 1.10 |
ENSMUST00000111045.9
|
Bdnf
|
brain derived neurotrophic factor |
| chr1_+_42992109 | 1.10 |
ENSMUST00000179766.3
|
Gpr45
|
G protein-coupled receptor 45 |
| chr5_-_92822984 | 1.09 |
ENSMUST00000060930.10
|
Ccdc158
|
coiled-coil domain containing 158 |
| chr6_-_33037191 | 1.08 |
ENSMUST00000066379.11
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr19_-_29790352 | 1.07 |
ENSMUST00000099525.5
|
Ranbp6
|
RAN binding protein 6 |
| chr11_+_76563281 | 1.07 |
ENSMUST00000056184.2
|
Bhlha9
|
basic helix-loop-helix family, member a9 |
| chr8_-_120904179 | 1.06 |
ENSMUST00000048786.13
|
Cibar2
|
CBY1 interacting BAR domain containing 2 |
| chr11_+_52123016 | 1.05 |
ENSMUST00000109072.2
|
Skp1
|
S-phase kinase-associated protein 1 |
| chr11_+_52122836 | 1.04 |
ENSMUST00000037324.12
ENSMUST00000166537.8 |
Skp1
|
S-phase kinase-associated protein 1 |
| chr11_+_121036969 | 1.04 |
ENSMUST00000039088.9
ENSMUST00000155694.2 |
Tex19.1
|
testis expressed gene 19.1 |
| chr4_-_135112956 | 1.03 |
ENSMUST00000105857.8
ENSMUST00000105858.8 ENSMUST00000064481.15 ENSMUST00000123632.2 |
Ncmap
|
noncompact myelin associated protein |
| chr9_-_75317233 | 1.02 |
ENSMUST00000049355.11
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr19_+_26725589 | 1.02 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_-_36484328 | 1.02 |
ENSMUST00000125304.8
ENSMUST00000115011.8 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr15_+_81943345 | 1.01 |
ENSMUST00000100396.4
|
4930407I10Rik
|
RIKEN cDNA 4930407I10 gene |
| chr2_+_80447389 | 1.00 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr13_-_111626562 | 1.00 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr1_+_153750081 | 1.00 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chr16_-_55755160 | 1.00 |
ENSMUST00000122280.8
ENSMUST00000121703.3 |
Cep97
|
centrosomal protein 97 |
| chr11_+_104576528 | 0.99 |
ENSMUST00000148007.3
ENSMUST00000212287.2 |
Gm11639
|
predicted gene 11639 |
| chr9_+_18203640 | 0.99 |
ENSMUST00000217031.2
|
Chordc1
|
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| chr19_+_6413703 | 0.99 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr14_-_66071337 | 0.98 |
ENSMUST00000225853.2
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_+_91178288 | 0.98 |
ENSMUST00000171112.8
ENSMUST00000191533.2 |
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chrX_+_70408507 | 0.98 |
ENSMUST00000132837.5
|
Mtmr1
|
myotubularin related protein 1 |
| chr11_+_21041291 | 0.97 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr10_+_127595590 | 0.97 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr12_-_103597663 | 0.96 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr4_+_129181407 | 0.95 |
ENSMUST00000102599.4
|
Sync
|
syncoilin |
| chr18_+_70605722 | 0.95 |
ENSMUST00000174118.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr7_+_30411634 | 0.93 |
ENSMUST00000005692.14
ENSMUST00000170371.2 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr10_+_28544356 | 0.93 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr10_-_39039790 | 0.93 |
ENSMUST00000076713.6
|
Ccn6
|
cellular communication network factor 6 |
| chr3_+_67337429 | 0.93 |
ENSMUST00000077271.9
|
Gfm1
|
G elongation factor, mitochondrial 1 |
| chr9_-_58448224 | 0.92 |
ENSMUST00000039788.11
|
Cd276
|
CD276 antigen |
| chr7_+_43837629 | 0.92 |
ENSMUST00000073713.8
|
Klk1b24
|
kallikrein 1-related peptidase b24 |
| chr1_+_164076822 | 0.92 |
ENSMUST00000169394.2
|
Slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr6_+_125026915 | 0.91 |
ENSMUST00000088294.12
ENSMUST00000032481.8 |
Acrbp
|
proacrosin binding protein |
| chr11_-_101169753 | 0.91 |
ENSMUST00000168089.2
ENSMUST00000017332.4 |
Coa3
|
cytochrome C oxidase assembly factor 3 |
| chr17_-_43003135 | 0.91 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr5_-_88823989 | 0.91 |
ENSMUST00000078945.12
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr13_+_81805941 | 0.90 |
ENSMUST00000049055.8
|
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr2_+_121287444 | 0.90 |
ENSMUST00000126764.2
|
Hypk
|
huntingtin interacting protein K |
| chrX_+_41238410 | 0.90 |
ENSMUST00000127618.8
|
Stag2
|
stromal antigen 2 |
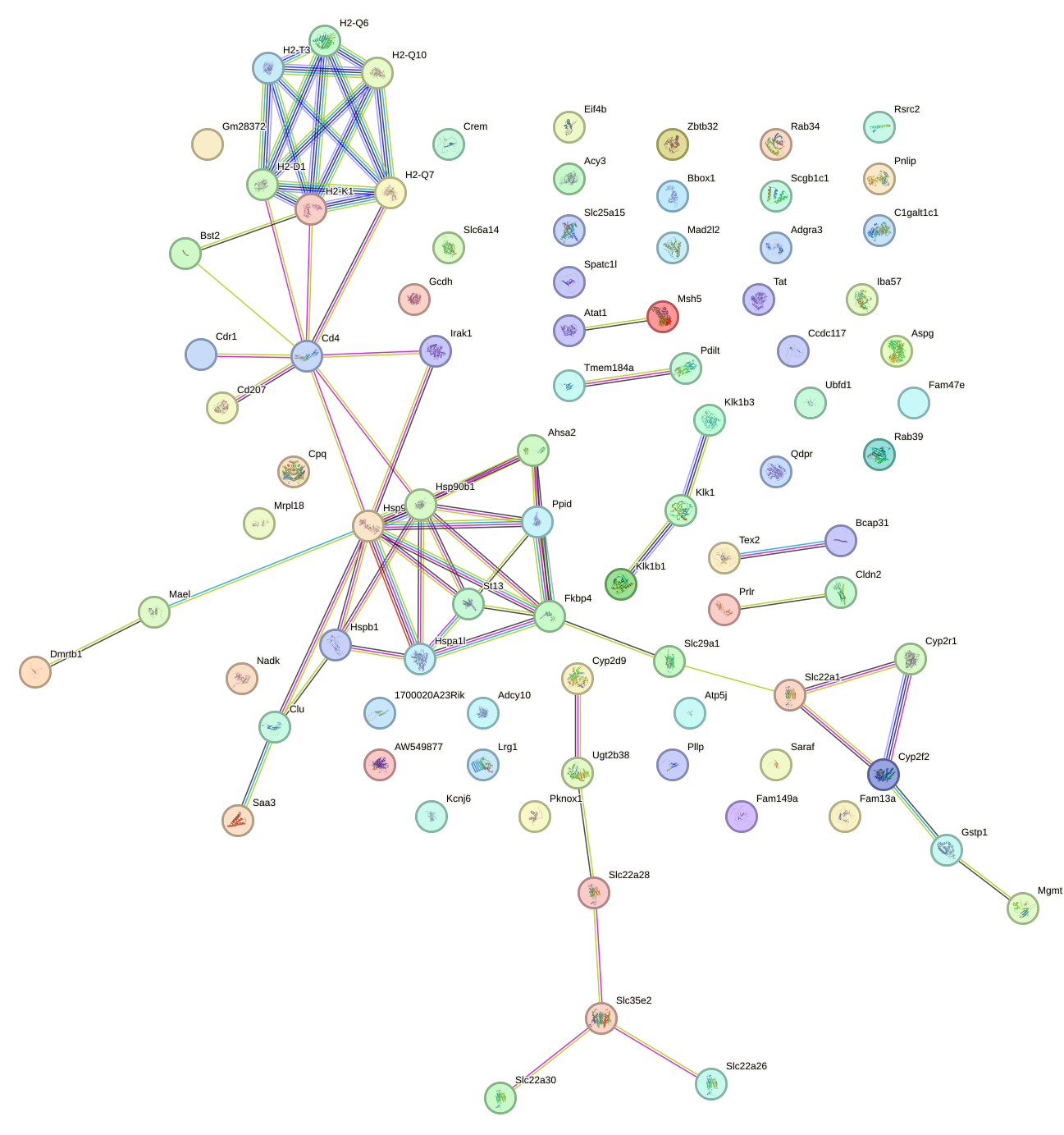
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 16.1 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 2.3 | 7.0 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 1.6 | 4.9 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 1.3 | 20.2 | GO:0015747 | urate transport(GO:0015747) |
| 1.3 | 12.9 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 1.2 | 3.6 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 1.1 | 16.6 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 1.1 | 3.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 1.1 | 3.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.1 | 7.5 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 1.0 | 3.9 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.9 | 2.8 | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000299) |
| 0.9 | 2.8 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.9 | 4.6 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.7 | 3.0 | GO:0039521 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) amino acid homeostasis(GO:0080144) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.7 | 5.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.7 | 4.7 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.6 | 5.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.6 | 3.6 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.6 | 1.8 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.6 | 3.5 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.6 | 5.3 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.5 | 1.6 | GO:0033128 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.5 | 3.0 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.5 | 3.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.5 | 1.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 2.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 5.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.4 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 2.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 1.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.4 | 2.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.4 | 6.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.4 | 1.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.4 | 2.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.4 | 1.4 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.3 | 1.0 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.3 | 3.7 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.3 | 15.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.3 | 1.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 1.2 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.3 | 5.8 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.3 | 0.9 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.3 | 0.6 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.3 | 0.9 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.3 | 2.7 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 1.9 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.3 | 0.8 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.3 | 1.5 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.3 | 0.8 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 | 0.5 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.2 | 5.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.7 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 1.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 0.9 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 0.6 | GO:1990918 | meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 1.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 1.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 0.6 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 1.2 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 1.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 1.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.2 | 1.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 0.9 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.2 | 2.9 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 0.7 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 2.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 0.7 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 1.3 | GO:0051036 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 2.1 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.2 | 1.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.4 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.6 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.7 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 4.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.7 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.7 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 3.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.9 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.4 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 1.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.9 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.9 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 1.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.0 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 5.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.7 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 1.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.4 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.8 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.3 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.3 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 0.4 | GO:0015755 | fructose transport(GO:0015755) |
| 0.1 | 0.3 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 2.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 4.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.6 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 2.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 2.7 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 1.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.4 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.8 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 1.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.9 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 2.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 0.4 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.1 | 1.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.9 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.6 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 2.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 4.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.6 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.9 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.2 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 3.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.9 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.8 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.5 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.7 | GO:1902579 | multi-organism transport(GO:0044766) multi-organism localization(GO:1902579) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:1902623 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 2.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.4 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.0 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.6 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 1.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 1.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.8 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.8 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.0 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.8 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.1 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.7 | GO:0090077 | foam cell differentiation(GO:0090077) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.4 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.5 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.7 | GO:0010524 | positive regulation of calcium ion transport into cytosol(GO:0010524) |
| 0.0 | 2.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.9 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.1 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.0 | 2.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 31.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.2 | 7.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 1.0 | 4.9 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.6 | 2.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.5 | 3.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.4 | 1.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.4 | 1.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.3 | 1.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.3 | 2.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 4.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 0.9 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.2 | 1.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.2 | 0.7 | GO:0060473 | cortical granule(GO:0060473) |
| 0.2 | 1.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 3.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 5.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.2 | 2.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 6.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 0.6 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.2 | 1.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 4.9 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 0.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 3.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 5.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 9.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 36.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 4.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 20.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.9 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 1.9 | 5.8 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.8 | 31.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 1.7 | 5.0 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.6 | 4.9 | GO:0035731 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 1.2 | 3.6 | GO:0004067 | asparaginase activity(GO:0004067) |
| 1.2 | 20.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.1 | 9.6 | GO:0032564 | dATP binding(GO:0032564) |
| 1.0 | 3.9 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.9 | 5.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.9 | 6.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.8 | 2.4 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.7 | 3.0 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.7 | 7.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.7 | 3.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.6 | 2.6 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.6 | 4.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.5 | 3.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.5 | 2.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 1.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.4 | 5.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 1.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 5.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 2.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 2.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 0.9 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.3 | 3.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.3 | 11.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.3 | 1.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 1.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 0.8 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.2 | 1.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.2 | 2.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 2.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.7 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.2 | 2.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 5.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.7 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.2 | 0.9 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 11.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 1.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 0.9 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.2 | 0.6 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 1.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 4.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 4.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 0.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.6 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.2 | 2.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 2.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 0.5 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.2 | 0.5 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.2 | 0.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 1.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 3.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 5.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 5.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 2.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 7.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 1.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 5.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 7.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 4.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.8 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 2.8 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.6 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 2.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 1.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 2.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 4.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 3.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.6 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.9 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 2.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.8 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 6.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 6.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 3.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 3.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.5 | 6.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.4 | 2.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.4 | 5.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 7.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 2.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 13.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 4.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 1.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 0.9 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 5.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 4.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 5.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 22.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 4.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 3.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 2.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.6 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |