Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
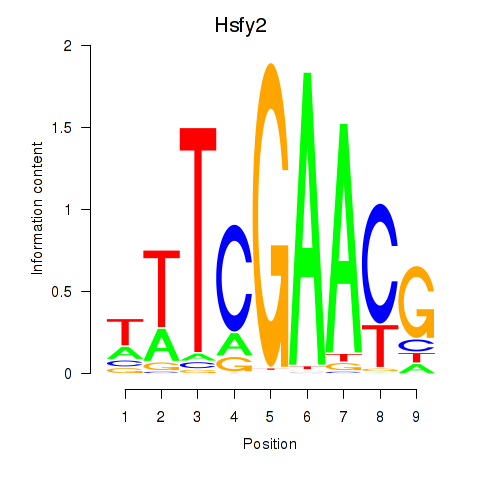
Results for Hsfy2
Z-value: 0.84
Transcription factors associated with Hsfy2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsfy2
|
ENSMUSG00000045336.6 | heat shock transcription factor, Y-linked 2 |
Activity profile of Hsfy2 motif
Sorted Z-values of Hsfy2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13467422 | 8.01 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr1_+_171246593 | 3.00 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr13_+_4484305 | 2.28 |
ENSMUST00000021630.15
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr16_+_22769822 | 2.13 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr3_+_142406827 | 2.01 |
ENSMUST00000044392.11
ENSMUST00000199519.5 |
Kyat3
|
kynurenine aminotransferase 3 |
| chr3_+_142406787 | 2.00 |
ENSMUST00000106218.8
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr18_+_87774402 | 1.98 |
ENSMUST00000091776.7
|
Gm5096
|
predicted gene 5096 |
| chr17_-_14914484 | 1.66 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2 |
| chr12_-_28673259 | 1.63 |
ENSMUST00000220836.2
|
Colec11
|
collectin sub-family member 11 |
| chr16_+_26400454 | 1.63 |
ENSMUST00000096129.9
ENSMUST00000166294.9 ENSMUST00000174202.8 ENSMUST00000023156.13 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr12_-_28673311 | 1.60 |
ENSMUST00000036136.9
|
Colec11
|
collectin sub-family member 11 |
| chr19_-_43512929 | 1.57 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr1_-_150341911 | 1.55 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr4_+_105014536 | 1.53 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr1_-_98023321 | 1.51 |
ENSMUST00000058762.15
ENSMUST00000097625.10 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr9_-_106124917 | 1.50 |
ENSMUST00000112524.9
ENSMUST00000219129.2 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr13_-_63036096 | 1.45 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr5_+_9163244 | 1.43 |
ENSMUST00000198935.2
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr1_+_45350698 | 1.43 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr2_-_10135449 | 1.40 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr15_+_55171138 | 1.31 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr4_+_108736350 | 1.25 |
ENSMUST00000106651.9
|
Rab3b
|
RAB3B, member RAS oncogene family |
| chr1_-_139487951 | 1.23 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr4_+_20007938 | 1.22 |
ENSMUST00000125799.8
ENSMUST00000121491.8 |
Ttpa
|
tocopherol (alpha) transfer protein |
| chr1_+_88093726 | 1.21 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr6_-_142517029 | 1.21 |
ENSMUST00000032374.9
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr9_+_37524966 | 1.20 |
ENSMUST00000215474.2
|
Siae
|
sialic acid acetylesterase |
| chr6_+_54241830 | 1.19 |
ENSMUST00000146114.8
|
Chn2
|
chimerin 2 |
| chr13_+_4099001 | 1.15 |
ENSMUST00000118717.10
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr2_-_32872032 | 1.15 |
ENSMUST00000195863.2
ENSMUST00000028129.13 |
Slc2a8
|
solute carrier family 2, (facilitated glucose transporter), member 8 |
| chr10_+_107107558 | 1.15 |
ENSMUST00000105280.5
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr3_-_131065658 | 1.14 |
ENSMUST00000029610.9
|
Hadh
|
hydroxyacyl-Coenzyme A dehydrogenase |
| chr1_+_132243849 | 1.09 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr4_+_108736260 | 1.08 |
ENSMUST00000106650.9
|
Rab3b
|
RAB3B, member RAS oncogene family |
| chr10_+_128030315 | 1.06 |
ENSMUST00000044776.13
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr11_-_75313350 | 1.00 |
ENSMUST00000125982.2
ENSMUST00000137103.8 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr5_-_108280406 | 0.99 |
ENSMUST00000119437.8
ENSMUST00000118036.8 ENSMUST00000061203.13 ENSMUST00000002837.11 |
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr10_-_112764879 | 0.98 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr2_+_155223728 | 0.98 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr6_-_113412708 | 0.96 |
ENSMUST00000032416.11
ENSMUST00000113089.8 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr14_+_66378382 | 0.94 |
ENSMUST00000022620.11
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr5_-_113163339 | 0.94 |
ENSMUST00000197776.2
ENSMUST00000065167.9 |
Grk3
|
G protein-coupled receptor kinase 3 |
| chr3_+_40905216 | 0.93 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr16_+_62674661 | 0.93 |
ENSMUST00000023629.9
|
Pros1
|
protein S (alpha) |
| chr2_-_32872019 | 0.91 |
ENSMUST00000153484.7
|
Slc2a8
|
solute carrier family 2, (facilitated glucose transporter), member 8 |
| chr8_+_95564949 | 0.89 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr9_+_44893077 | 0.89 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chrX_+_10118600 | 0.87 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr16_-_18880821 | 0.85 |
ENSMUST00000200568.2
|
Iglc1
|
immunoglobulin lambda constant 1 |
| chr18_+_84869883 | 0.84 |
ENSMUST00000163083.2
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr10_+_62897353 | 0.83 |
ENSMUST00000178684.3
ENSMUST00000020266.15 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr13_+_3684032 | 0.83 |
ENSMUST00000042288.8
|
Asb13
|
ankyrin repeat and SOCS box-containing 13 |
| chr11_-_117764258 | 0.82 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr9_-_55419442 | 0.81 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr13_-_48779072 | 0.81 |
ENSMUST00000035824.11
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr10_-_81121596 | 0.80 |
ENSMUST00000218484.2
|
Tjp3
|
tight junction protein 3 |
| chr8_-_41507808 | 0.80 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr3_+_137983250 | 0.78 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr9_-_50466470 | 0.78 |
ENSMUST00000119103.2
|
Bco2
|
beta-carotene oxygenase 2 |
| chr10_+_128030500 | 0.77 |
ENSMUST00000123291.2
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chrX_-_142610371 | 0.77 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chr6_-_124718316 | 0.76 |
ENSMUST00000004389.6
|
Grcc10
|
gene rich cluster, C10 gene |
| chr9_+_50466127 | 0.76 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr6_-_98319684 | 0.75 |
ENSMUST00000164491.3
|
Mdfic2
|
MyoD family inhibitor domain containing 2 |
| chr11_-_46581135 | 0.73 |
ENSMUST00000169584.8
|
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr11_-_69553390 | 0.71 |
ENSMUST00000129224.8
ENSMUST00000155200.8 |
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr11_+_60619224 | 0.68 |
ENSMUST00000018743.5
|
Mief2
|
mitochondrial elongation factor 2 |
| chr10_-_128204545 | 0.66 |
ENSMUST00000220027.2
|
Coq10a
|
coenzyme Q10A |
| chr8_+_46080840 | 0.66 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr9_-_75317233 | 0.66 |
ENSMUST00000049355.11
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr10_-_128204806 | 0.66 |
ENSMUST00000043211.7
ENSMUST00000220227.2 |
Coq10a
|
coenzyme Q10A |
| chr11_-_69553451 | 0.65 |
ENSMUST00000018905.12
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr4_+_41465134 | 0.64 |
ENSMUST00000030154.7
|
Nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr6_+_48514518 | 0.64 |
ENSMUST00000040361.8
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2 |
| chr13_+_38335232 | 0.63 |
ENSMUST00000124830.3
|
Dsp
|
desmoplakin |
| chr1_-_72323464 | 0.61 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chrM_-_14061 | 0.61 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chrX_+_72386220 | 0.61 |
ENSMUST00000114499.8
ENSMUST00000033731.4 |
Zfp275
|
zinc finger protein 275 |
| chr5_+_21577640 | 0.61 |
ENSMUST00000035799.6
|
Fgl2
|
fibrinogen-like protein 2 |
| chr11_-_76737794 | 0.61 |
ENSMUST00000021201.6
|
Cpd
|
carboxypeptidase D |
| chr4_+_102446883 | 0.57 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr19_-_10859087 | 0.57 |
ENSMUST00000144681.2
|
Tmem109
|
transmembrane protein 109 |
| chr8_+_46428551 | 0.57 |
ENSMUST00000034051.7
ENSMUST00000150943.2 |
Ufsp2
|
UFM1-specific peptidase 2 |
| chr6_+_48514578 | 0.56 |
ENSMUST00000203011.2
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2 |
| chr11_-_116089866 | 0.56 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr17_-_66258110 | 0.56 |
ENSMUST00000233580.2
ENSMUST00000024906.6 |
Twsg1
|
twisted gastrulation BMP signaling modulator 1 |
| chr10_-_61814852 | 0.56 |
ENSMUST00000105453.8
ENSMUST00000105452.9 ENSMUST00000105454.3 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr2_-_80277969 | 0.55 |
ENSMUST00000028389.4
|
Frzb
|
frizzled-related protein |
| chr11_+_74540284 | 0.53 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr6_-_48817675 | 0.53 |
ENSMUST00000203265.3
ENSMUST00000205159.3 |
Tmem176b
|
transmembrane protein 176B |
| chr18_+_70058533 | 0.52 |
ENSMUST00000043929.11
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr2_+_32496957 | 0.52 |
ENSMUST00000113290.8
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr19_-_38113056 | 0.52 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr8_-_78337226 | 0.51 |
ENSMUST00000034030.15
|
Tmem184c
|
transmembrane protein 184C |
| chr5_+_8943943 | 0.50 |
ENSMUST00000196067.2
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr16_+_35758836 | 0.50 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr12_-_98700886 | 0.50 |
ENSMUST00000085116.4
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr11_+_66915969 | 0.49 |
ENSMUST00000079077.12
ENSMUST00000061786.6 |
Tmem220
|
transmembrane protein 220 |
| chr14_+_29730931 | 0.49 |
ENSMUST00000067620.12
|
Chdh
|
choline dehydrogenase |
| chr6_-_5256282 | 0.48 |
ENSMUST00000031773.9
|
Pon3
|
paraoxonase 3 |
| chr11_-_3402355 | 0.47 |
ENSMUST00000077078.12
ENSMUST00000064364.3 |
Rnf185
|
ring finger protein 185 |
| chr17_-_46985207 | 0.47 |
ENSMUST00000232697.2
ENSMUST00000233136.2 |
Rrp36
|
ribosomal RNA processing 36 |
| chr1_-_119576632 | 0.47 |
ENSMUST00000163147.8
ENSMUST00000052404.13 ENSMUST00000191046.7 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr2_+_32496990 | 0.47 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr9_+_44045859 | 0.46 |
ENSMUST00000034650.15
ENSMUST00000098852.3 ENSMUST00000216002.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr17_-_46985181 | 0.46 |
ENSMUST00000024766.7
|
Rrp36
|
ribosomal RNA processing 36 |
| chr7_-_44711130 | 0.46 |
ENSMUST00000211337.2
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chrX_-_139501246 | 0.46 |
ENSMUST00000112996.9
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr8_+_46080746 | 0.46 |
ENSMUST00000145458.9
ENSMUST00000134321.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr5_+_104607316 | 0.45 |
ENSMUST00000086831.4
|
Pkd2
|
polycystin 2, transient receptor potential cation channel |
| chr7_+_48896560 | 0.45 |
ENSMUST00000184945.8
|
Nav2
|
neuron navigator 2 |
| chr7_-_44711075 | 0.45 |
ENSMUST00000007981.9
ENSMUST00000210500.2 ENSMUST00000210493.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr11_-_120464062 | 0.44 |
ENSMUST00000026122.11
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr11_-_120463667 | 0.44 |
ENSMUST00000168360.2
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr1_+_57813759 | 0.44 |
ENSMUST00000167971.8
ENSMUST00000170139.8 ENSMUST00000171699.8 ENSMUST00000164302.8 |
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr4_+_98284054 | 0.43 |
ENSMUST00000107033.8
ENSMUST00000107034.8 |
Patj
|
PATJ, crumbs cell polarity complex component |
| chr16_+_16031182 | 0.42 |
ENSMUST00000039408.3
|
Pkp2
|
plakophilin 2 |
| chr11_+_95734419 | 0.42 |
ENSMUST00000107708.2
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr5_-_108280350 | 0.42 |
ENSMUST00000119784.2
ENSMUST00000117759.2 |
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr11_-_50216426 | 0.42 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr8_+_53964721 | 0.42 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr15_-_89314054 | 0.41 |
ENSMUST00000023289.13
|
Chkb
|
choline kinase beta |
| chr9_+_57037974 | 0.41 |
ENSMUST00000160147.8
ENSMUST00000161663.8 ENSMUST00000034836.16 ENSMUST00000161182.8 |
Man2c1
|
mannosidase, alpha, class 2C, member 1 |
| chr10_-_75670217 | 0.40 |
ENSMUST00000218500.2
|
Gstt2
|
glutathione S-transferase, theta 2 |
| chr11_+_95734028 | 0.40 |
ENSMUST00000107709.8
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr10_-_127502541 | 0.40 |
ENSMUST00000026469.9
|
Nab2
|
Ngfi-A binding protein 2 |
| chr6_-_48817389 | 0.39 |
ENSMUST00000204783.3
|
Tmem176b
|
transmembrane protein 176B |
| chr10_-_89568106 | 0.39 |
ENSMUST00000020109.5
|
Actr6
|
ARP6 actin-related protein 6 |
| chr2_-_101479846 | 0.39 |
ENSMUST00000078494.6
ENSMUST00000160722.8 ENSMUST00000160037.8 |
Rag1
Iftap
|
recombination activating 1 intraflagellar transport associated protein |
| chr10_-_127502467 | 0.38 |
ENSMUST00000099157.4
|
Nab2
|
Ngfi-A binding protein 2 |
| chr2_+_101716577 | 0.38 |
ENSMUST00000028584.8
|
Commd9
|
COMM domain containing 9 |
| chr7_+_64042393 | 0.38 |
ENSMUST00000037205.11
|
Mcee
|
methylmalonyl CoA epimerase |
| chr6_-_116605914 | 0.38 |
ENSMUST00000079749.6
|
Zfp422
|
zinc finger protein 422 |
| chr3_-_104960264 | 0.38 |
ENSMUST00000098763.7
ENSMUST00000197437.5 |
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr15_+_54609011 | 0.37 |
ENSMUST00000050027.9
|
Ccn3
|
cellular communication network factor 3 |
| chr12_-_72283465 | 0.37 |
ENSMUST00000021497.16
ENSMUST00000137990.2 |
Rtn1
|
reticulon 1 |
| chr6_-_5256225 | 0.37 |
ENSMUST00000125686.8
|
Pon3
|
paraoxonase 3 |
| chr7_+_64042514 | 0.37 |
ENSMUST00000206194.2
|
Mcee
|
methylmalonyl CoA epimerase |
| chr5_+_53424471 | 0.37 |
ENSMUST00000147148.5
|
Smim20
|
small integral membrane protein 20 |
| chr13_+_37529184 | 0.37 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr7_+_64042482 | 0.37 |
ENSMUST00000206882.2
|
Mcee
|
methylmalonyl CoA epimerase |
| chr19_-_10859046 | 0.36 |
ENSMUST00000128835.8
|
Tmem109
|
transmembrane protein 109 |
| chr8_-_78337297 | 0.36 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chr10_-_59452489 | 0.36 |
ENSMUST00000020312.13
|
Mcu
|
mitochondrial calcium uniporter |
| chr2_+_26899935 | 0.36 |
ENSMUST00000114005.9
ENSMUST00000114004.8 ENSMUST00000114006.8 ENSMUST00000114007.8 ENSMUST00000133807.2 |
Cacfd1
|
calcium channel flower domain containing 1 |
| chr9_+_95441652 | 0.35 |
ENSMUST00000079597.7
|
Paqr9
|
progestin and adipoQ receptor family member IX |
| chr6_+_34361153 | 0.35 |
ENSMUST00000038383.14
ENSMUST00000115051.8 |
Akr1b10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr13_-_91372072 | 0.35 |
ENSMUST00000022119.6
|
Atg10
|
autophagy related 10 |
| chr1_+_13738967 | 0.34 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr14_+_60615690 | 0.32 |
ENSMUST00000239079.2
|
Amer2
|
APC membrane recruitment 2 |
| chr9_+_95739650 | 0.32 |
ENSMUST00000034980.9
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr15_-_80148477 | 0.32 |
ENSMUST00000052499.9
|
Rps19bp1
|
ribosomal protein S19 binding protein 1 |
| chr6_-_116605882 | 0.31 |
ENSMUST00000057540.6
|
Zfp422
|
zinc finger protein 422 |
| chr7_+_101312840 | 0.31 |
ENSMUST00000001884.14
|
Clpb
|
ClpB caseinolytic peptidase B |
| chr11_+_70910418 | 0.31 |
ENSMUST00000133413.8
ENSMUST00000164220.8 ENSMUST00000048807.12 |
Mis12
|
MIS12 kinetochore complex component |
| chr3_+_7568481 | 0.31 |
ENSMUST00000051064.9
ENSMUST00000193010.2 |
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr16_-_31767250 | 0.31 |
ENSMUST00000202722.2
|
0610012G03Rik
|
RIKEN cDNA 0610012G03 gene |
| chr14_-_55019387 | 0.31 |
ENSMUST00000022787.8
|
Slc7a8
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 |
| chr7_-_44803859 | 0.31 |
ENSMUST00000210125.2
|
Aldh16a1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr8_+_89309408 | 0.31 |
ENSMUST00000211113.2
|
Nkd1
|
naked cuticle 1 |
| chrM_+_14138 | 0.30 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr5_+_77099154 | 0.30 |
ENSMUST00000031160.16
ENSMUST00000120912.8 ENSMUST00000117536.8 |
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chrX_-_95000496 | 0.29 |
ENSMUST00000079987.13
ENSMUST00000113864.3 |
Las1l
|
LAS1-like (S. cerevisiae) |
| chr8_-_4375320 | 0.29 |
ENSMUST00000098950.6
|
Elavl1
|
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
| chrX_+_99019176 | 0.29 |
ENSMUST00000113781.8
ENSMUST00000113783.8 ENSMUST00000113779.8 ENSMUST00000113776.8 ENSMUST00000113775.8 ENSMUST00000113780.8 ENSMUST00000113778.8 ENSMUST00000113777.8 ENSMUST00000071453.3 |
Eda
|
ectodysplasin-A |
| chr19_-_29790352 | 0.29 |
ENSMUST00000099525.5
|
Ranbp6
|
RAN binding protein 6 |
| chr3_-_104960437 | 0.29 |
ENSMUST00000077548.12
|
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chrX_-_103457431 | 0.29 |
ENSMUST00000033695.6
|
Abcb7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr5_+_31652079 | 0.28 |
ENSMUST00000076949.13
ENSMUST00000202394.4 |
Gpn1
|
GPN-loop GTPase 1 |
| chrM_+_2743 | 0.28 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr2_+_76505619 | 0.28 |
ENSMUST00000111920.2
|
Plekha3
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 3 |
| chr18_-_37777238 | 0.28 |
ENSMUST00000066272.6
|
Taf7
|
TATA-box binding protein associated factor 7 |
| chr1_-_93729650 | 0.27 |
ENSMUST00000027503.14
|
Dtymk
|
deoxythymidylate kinase |
| chr8_+_107141948 | 0.27 |
ENSMUST00000034382.8
ENSMUST00000212606.2 |
Zfp90
|
zinc finger protein 90 |
| chr7_+_101312912 | 0.27 |
ENSMUST00000106998.8
ENSMUST00000209579.2 |
Clpb
|
ClpB caseinolytic peptidase B |
| chr17_+_87943401 | 0.26 |
ENSMUST00000235125.2
ENSMUST00000053577.9 ENSMUST00000234009.2 |
Epcam
|
epithelial cell adhesion molecule |
| chr1_+_57814001 | 0.26 |
ENSMUST00000167085.8
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr13_+_42862957 | 0.26 |
ENSMUST00000066928.12
ENSMUST00000148891.8 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr1_-_190795092 | 0.26 |
ENSMUST00000085633.12
ENSMUST00000110891.8 ENSMUST00000027945.6 ENSMUST00000110893.10 |
Tatdn3
|
TatD DNase domain containing 3 |
| chr14_-_75185281 | 0.26 |
ENSMUST00000088970.7
ENSMUST00000228252.2 |
Lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr6_+_123206802 | 0.26 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr7_+_48608800 | 0.26 |
ENSMUST00000183659.8
|
Nav2
|
neuron navigator 2 |
| chr3_-_113263974 | 0.26 |
ENSMUST00000098667.5
|
Amy2a2
|
amylase 2a2 |
| chrX_-_56438380 | 0.25 |
ENSMUST00000143310.2
ENSMUST00000098470.9 ENSMUST00000114726.8 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr4_-_14826587 | 0.25 |
ENSMUST00000117268.9
ENSMUST00000236953.2 |
Otud6b
|
OTU domain containing 6B |
| chr14_-_55881257 | 0.25 |
ENSMUST00000122358.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr5_+_135178509 | 0.25 |
ENSMUST00000153183.8
|
Tbl2
|
transducin (beta)-like 2 |
| chr6_+_113615428 | 0.24 |
ENSMUST00000059286.14
ENSMUST00000089023.11 ENSMUST00000089022.9 |
Irak2
|
interleukin-1 receptor-associated kinase 2 |
| chr13_+_63963054 | 0.24 |
ENSMUST00000021926.13
ENSMUST00000067821.13 ENSMUST00000144763.2 ENSMUST00000021925.14 ENSMUST00000238465.2 |
Ercc6l2
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 like 2 |
| chr7_-_66038033 | 0.24 |
ENSMUST00000015277.14
|
Lrrk1
|
leucine-rich repeat kinase 1 |
| chr14_-_55880980 | 0.24 |
ENSMUST00000132338.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr7_+_12568647 | 0.24 |
ENSMUST00000004614.15
|
Zfp110
|
zinc finger protein 110 |
| chr8_-_22675773 | 0.24 |
ENSMUST00000046916.9
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr1_+_163607143 | 0.24 |
ENSMUST00000077642.12
ENSMUST00000027877.7 |
Kifap3
|
kinesin-associated protein 3 |
| chr9_+_95739747 | 0.23 |
ENSMUST00000215311.2
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr7_-_78432774 | 0.23 |
ENSMUST00000032841.7
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr5_-_90514360 | 0.23 |
ENSMUST00000168058.7
ENSMUST00000014421.15 |
Ankrd17
|
ankyrin repeat domain 17 |
| chr7_+_44498640 | 0.23 |
ENSMUST00000054343.15
ENSMUST00000142880.3 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr11_-_82882613 | 0.23 |
ENSMUST00000092840.11
ENSMUST00000038211.13 |
Slfn9
|
schlafen 9 |
| chr14_+_26300693 | 0.23 |
ENSMUST00000203874.3
ENSMUST00000037585.9 |
Dennd6a
|
DENN/MADD domain containing 6A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsfy2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.8 | 2.3 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.7 | 2.1 | GO:0097037 | heme export(GO:0097037) |
| 0.5 | 1.6 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.5 | 2.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.4 | 1.1 | GO:0046491 | L-methylmalonyl-CoA metabolic process(GO:0046491) |
| 0.4 | 1.5 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 1.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.3 | 1.5 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.3 | 1.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 0.9 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.3 | 1.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 0.8 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.3 | 4.0 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.2 | 0.8 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 0.2 | 0.6 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.2 | 0.8 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 1.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.8 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.2 | 1.5 | GO:0031179 | peptide modification(GO:0031179) |
| 0.2 | 1.6 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 0.5 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.2 | 0.8 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 0.8 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 2.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.3 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.5 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 1.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 1.6 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.4 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 1.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.9 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 1.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.5 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.6 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.1 | 0.5 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 2.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 1.2 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 | 1.0 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.5 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.2 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.6 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.9 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 1.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.8 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.3 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 | 0.8 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.2 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.1 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.9 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 0.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) detection of oxygen(GO:0003032) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.7 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.1 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.0 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.2 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.0 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.9 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.5 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 1.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 2.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) sulfur oxidation(GO:0019417) |
| 0.0 | 0.7 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.3 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.7 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.2 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.8 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 5.8 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.0 | 0.2 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 1.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.6 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.2 | 0.7 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.2 | 0.8 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 0.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 0.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 1.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 1.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.5 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.7 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.2 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 3.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.8 | 4.0 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.8 | 2.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 1.6 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.5 | 1.5 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.4 | 1.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.4 | 1.6 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.4 | 1.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.4 | 1.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.4 | 1.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.4 | 1.1 | GO:0004493 | methylmalonyl-CoA epimerase activity(GO:0004493) |
| 0.3 | 1.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 0.8 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 0.9 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 0.8 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.2 | 0.8 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 1.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.3 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.2 | 0.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 1.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 1.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.7 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.1 | 0.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 3.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 8.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.3 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.4 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.3 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.8 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 0.5 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 1.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 2.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.5 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.6 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.9 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.9 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 4.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.4 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 3.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |