Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
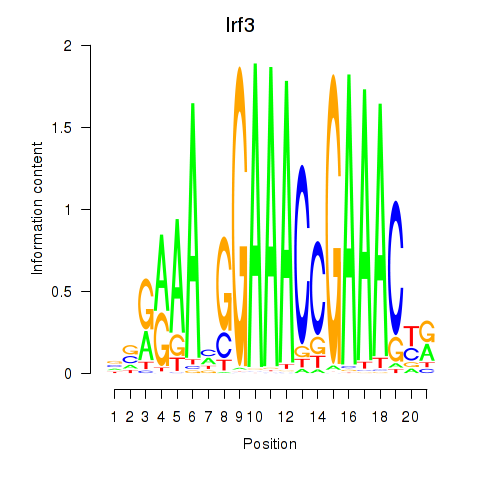
Results for Irf3
Z-value: 1.18
Transcription factors associated with Irf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf3
|
ENSMUSG00000003184.16 | interferon regulatory factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf3 | mm39_v1_chr7_+_44647072_44647126 | 0.24 | 1.6e-01 | Click! |
Activity profile of Irf3 motif
Sorted Z-values of Irf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_48883053 | 7.01 |
ENSMUST00000059930.3
ENSMUST00000068063.4 |
Gm12185
Tgtp1
|
predicted gene 12185 T cell specific GTPase 1 |
| chr11_-_48955028 | 6.35 |
ENSMUST00000046745.7
|
Tgtp2
|
T cell specific GTPase 2 |
| chr4_-_156285247 | 5.48 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr19_+_34560922 | 4.46 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr2_-_173060647 | 3.82 |
ENSMUST00000109116.3
ENSMUST00000029018.14 |
Zbp1
|
Z-DNA binding protein 1 |
| chr5_+_115034997 | 3.49 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr7_-_140846328 | 3.37 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chr5_-_121045568 | 3.32 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr11_+_72192455 | 2.97 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chr17_+_34406762 | 2.90 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr17_+_34406523 | 2.88 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_+_58090382 | 2.78 |
ENSMUST00000035266.11
ENSMUST00000094169.11 ENSMUST00000168280.2 ENSMUST00000058704.9 |
Igtp
Irgm2
|
interferon gamma induced GTPase immunity-related GTPase family M member 2 |
| chr5_-_121025645 | 2.64 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr11_-_48762170 | 2.52 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr16_+_23428650 | 2.49 |
ENSMUST00000038423.6
ENSMUST00000211349.2 |
Rtp4
|
receptor transporter protein 4 |
| chr11_-_100595019 | 2.44 |
ENSMUST00000017974.13
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr15_-_77295234 | 2.13 |
ENSMUST00000089452.6
ENSMUST00000081776.11 |
Apol9a
|
apolipoprotein L 9a |
| chr3_+_142236086 | 2.09 |
ENSMUST00000171263.8
ENSMUST00000045097.11 |
Gbp7
|
guanylate binding protein 7 |
| chr15_+_77613239 | 1.86 |
ENSMUST00000230979.2
ENSMUST00000109775.4 |
Apol9b
|
apolipoprotein L 9b |
| chr5_-_120950570 | 1.76 |
ENSMUST00000117193.8
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr11_-_83421333 | 1.76 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr5_+_120950690 | 1.42 |
ENSMUST00000086377.9
|
Oas1b
|
2'-5' oligoadenylate synthetase 1B |
| chr8_-_71990085 | 1.36 |
ENSMUST00000051672.9
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr10_+_128106414 | 1.35 |
ENSMUST00000085708.3
ENSMUST00000105238.10 |
Stat2
|
signal transducer and activator of transcription 2 |
| chr18_-_32692967 | 1.32 |
ENSMUST00000174000.2
ENSMUST00000174459.2 |
Gypc
|
glycophorin C |
| chr17_-_36331416 | 1.28 |
ENSMUST00000174063.2
ENSMUST00000113760.10 |
H2-T24
|
histocompatibility 2, T region locus 24 |
| chr15_+_77613348 | 1.24 |
ENSMUST00000230742.2
|
Apol9b
|
apolipoprotein L 9b |
| chr14_+_55815879 | 1.22 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr3_+_142265787 | 1.22 |
ENSMUST00000199325.5
ENSMUST00000106222.9 |
Gbp3
|
guanylate binding protein 3 |
| chr14_+_55815817 | 1.21 |
ENSMUST00000174259.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr14_+_55815580 | 1.17 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr17_-_34406193 | 1.15 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr14_+_55815999 | 1.14 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr11_+_119283887 | 1.12 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr16_-_35691914 | 1.11 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr6_-_39095144 | 1.08 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr7_-_103964662 | 1.06 |
ENSMUST00000106837.8
ENSMUST00000106839.9 ENSMUST00000070943.7 |
Trim12a
|
tripartite motif-containing 12A |
| chr8_-_124045292 | 1.01 |
ENSMUST00000118395.2
ENSMUST00000035495.15 |
Fanca
|
Fanconi anemia, complementation group A |
| chr1_-_85526517 | 0.94 |
ENSMUST00000093508.7
|
Sp110
|
Sp110 nuclear body protein |
| chr2_+_180231038 | 0.85 |
ENSMUST00000029087.4
|
Ogfr
|
opioid growth factor receptor |
| chr7_-_102214636 | 0.83 |
ENSMUST00000106913.3
ENSMUST00000033264.12 |
Trim21
|
tripartite motif-containing 21 |
| chr3_+_142265904 | 0.79 |
ENSMUST00000128609.8
ENSMUST00000029935.14 |
Gbp3
|
guanylate binding protein 3 |
| chr7_+_78563964 | 0.73 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr1_-_75294234 | 0.67 |
ENSMUST00000066668.14
ENSMUST00000185797.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr3_+_142266113 | 0.66 |
ENSMUST00000106221.8
|
Gbp3
|
guanylate binding protein 3 |
| chr1_+_85528392 | 0.66 |
ENSMUST00000080204.11
|
Sp140
|
Sp140 nuclear body protein |
| chr7_+_78563184 | 0.64 |
ENSMUST00000121645.8
|
Isg20
|
interferon-stimulated protein |
| chr16_-_10603389 | 0.59 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr2_-_166904902 | 0.57 |
ENSMUST00000048988.14
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr1_-_75293793 | 0.55 |
ENSMUST00000187836.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr11_+_109253598 | 0.55 |
ENSMUST00000106702.4
ENSMUST00000020930.14 |
Gna13
|
guanine nucleotide binding protein, alpha 13 |
| chr2_-_166904625 | 0.55 |
ENSMUST00000128676.2
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr15_-_82128538 | 0.54 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr7_+_78564062 | 0.48 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr3_-_151455514 | 0.48 |
ENSMUST00000029671.9
|
Ifi44
|
interferon-induced protein 44 |
| chr6_-_125208738 | 0.46 |
ENSMUST00000043422.8
|
Tapbpl
|
TAP binding protein-like |
| chr7_+_78563513 | 0.46 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr2_-_152174565 | 0.44 |
ENSMUST00000028964.14
|
Rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr2_-_155668567 | 0.43 |
ENSMUST00000109638.2
ENSMUST00000134278.2 |
Eif6
|
eukaryotic translation initiation factor 6 |
| chr6_-_34294377 | 0.42 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr1_-_75293637 | 0.39 |
ENSMUST00000185419.7
ENSMUST00000187000.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr7_+_103893658 | 0.38 |
ENSMUST00000106849.9
ENSMUST00000060315.12 |
Trim34a
|
tripartite motif-containing 34A |
| chr1_-_75293942 | 0.36 |
ENSMUST00000187075.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr2_-_152174273 | 0.31 |
ENSMUST00000109847.9
|
Rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr1_-_75293589 | 0.30 |
ENSMUST00000113605.10
|
Dnpep
|
aspartyl aminopeptidase |
| chr13_+_42205790 | 0.30 |
ENSMUST00000220525.2
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr1_-_75293447 | 0.28 |
ENSMUST00000189551.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr14_-_55828511 | 0.27 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr16_+_35759346 | 0.26 |
ENSMUST00000023622.13
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr10_-_81332163 | 0.25 |
ENSMUST00000020463.14
|
Ncln
|
nicalin |
| chr9_+_20779924 | 0.24 |
ENSMUST00000043911.8
|
Shfl
|
shiftless antiviral inhibitor of ribosomal frameshifting |
| chr9_-_35087348 | 0.24 |
ENSMUST00000119847.2
ENSMUST00000034539.12 |
Dcps
|
decapping enzyme, scavenger |
| chr5_-_120950549 | 0.24 |
ENSMUST00000125547.2
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr6_-_54949587 | 0.24 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr6_+_34840151 | 0.22 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr7_-_3551003 | 0.22 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr10_-_81332147 | 0.21 |
ENSMUST00000118498.8
|
Ncln
|
nicalin |
| chr7_-_104002534 | 0.21 |
ENSMUST00000130139.3
ENSMUST00000059037.15 |
Trim12c
|
tripartite motif-containing 12C |
| chr4_-_40239778 | 0.19 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr8_-_106665060 | 0.18 |
ENSMUST00000034369.10
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10 |
| chr6_+_34840057 | 0.17 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr7_+_128125339 | 0.16 |
ENSMUST00000033136.9
|
Bag3
|
BCL2-associated athanogene 3 |
| chr2_-_62476487 | 0.15 |
ENSMUST00000112459.4
ENSMUST00000028259.12 |
Ifih1
|
interferon induced with helicase C domain 1 |
| chr1_-_75293997 | 0.14 |
ENSMUST00000189282.3
ENSMUST00000191254.7 |
Dnpep
|
aspartyl aminopeptidase |
| chrX_+_100473161 | 0.14 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr12_-_83643964 | 0.09 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr17_+_57665691 | 0.08 |
ENSMUST00000086763.13
ENSMUST00000233376.2 ENSMUST00000233840.2 ENSMUST00000232808.2 ENSMUST00000004850.8 |
Adgre1
|
adhesion G protein-coupled receptor E1 |
| chr9_+_108385247 | 0.06 |
ENSMUST00000207810.2
ENSMUST00000207862.2 ENSMUST00000208581.2 ENSMUST00000134939.9 ENSMUST00000207713.2 |
Qars
|
glutaminyl-tRNA synthetase |
| chr19_+_6096606 | 0.06 |
ENSMUST00000138532.8
ENSMUST00000129081.8 ENSMUST00000156550.8 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr1_-_85087567 | 0.05 |
ENSMUST00000161675.3
ENSMUST00000160792.8 |
A530032D15Rik
|
RIKEN cDNA A530032D15Rik gene |
| chr7_-_15656285 | 0.04 |
ENSMUST00000044355.11
|
Selenow
|
selenoprotein W |
| chr6_-_3399451 | 0.04 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr7_+_103893672 | 0.03 |
ENSMUST00000106848.8
|
Trim34a
|
tripartite motif-containing 34A |
| chr7_-_100164007 | 0.02 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr17_+_37148015 | 0.02 |
ENSMUST00000179968.8
ENSMUST00000130367.8 ENSMUST00000053434.15 ENSMUST00000130801.8 ENSMUST00000144182.8 ENSMUST00000123715.8 |
Trim26
|
tripartite motif-containing 26 |
| chr2_-_173117936 | 0.01 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr15_-_77417512 | 0.01 |
ENSMUST00000062562.7
ENSMUST00000230863.2 |
Apol7c
|
apolipoprotein L 7c |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.9 | 2.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.7 | 5.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.6 | 3.4 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.4 | 23.0 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.4 | 1.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 2.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.4 | 2.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 4.3 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.3 | 3.8 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.3 | 1.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.4 | GO:0018931 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.1 | 2.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 4.3 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.1 | 0.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 2.1 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.6 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.1 | 1.4 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.2 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.1 | 0.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.2 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 1.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 4.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 2.1 | GO:0048525 | negative regulation of viral process(GO:0048525) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 1.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.7 | 5.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.5 | 7.5 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.2 | 1.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 2.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 0.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 12.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) omegasome membrane(GO:1903349) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 1.3 | 3.8 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 1.0 | 11.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.6 | 1.8 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 2.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.5 | 5.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 2.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.3 | 5.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 2.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 1.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 18.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 1.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 2.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 5.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 3.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 20.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 4.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.8 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 5.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 2.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.6 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.5 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |