Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
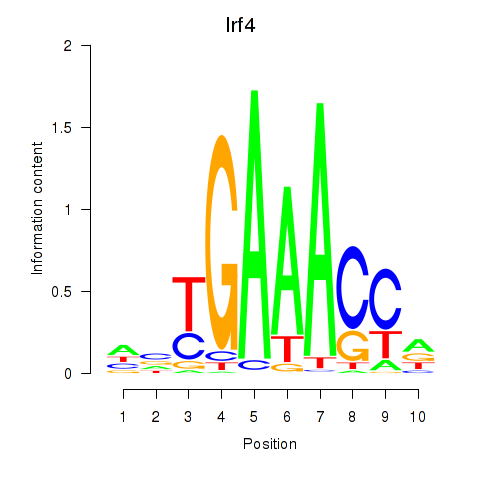
Results for Irf4
Z-value: 0.75
Transcription factors associated with Irf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf4
|
ENSMUSG00000021356.11 | interferon regulatory factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf4 | mm39_v1_chr13_+_30933209_30933290 | 0.10 | 5.7e-01 | Click! |
Activity profile of Irf4 motif
Sorted Z-values of Irf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_115353290 | 3.40 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr9_-_65330231 | 2.43 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr2_-_173060647 | 2.36 |
ENSMUST00000109116.3
ENSMUST00000029018.14 |
Zbp1
|
Z-DNA binding protein 1 |
| chr19_-_58443012 | 2.25 |
ENSMUST00000129100.8
ENSMUST00000123957.2 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr17_-_35081456 | 1.95 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr9_+_44953723 | 1.87 |
ENSMUST00000034600.5
|
Mpzl2
|
myelin protein zero-like 2 |
| chr14_+_14475188 | 1.84 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr6_+_82018604 | 1.83 |
ENSMUST00000042974.15
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr1_-_97904958 | 1.66 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr5_-_104169696 | 1.60 |
ENSMUST00000119025.2
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr4_-_19922599 | 1.59 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr19_-_58442866 | 1.58 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr5_-_104169785 | 1.55 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr17_+_35481702 | 1.46 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr4_-_46536088 | 1.44 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr1_-_164763091 | 1.37 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr11_+_48977852 | 1.36 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr19_-_58443830 | 1.34 |
ENSMUST00000026076.14
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr17_-_35081129 | 1.30 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr19_-_58443593 | 1.28 |
ENSMUST00000135730.2
ENSMUST00000152507.8 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr11_+_46701619 | 1.28 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr7_-_65020655 | 1.24 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr9_+_14187597 | 1.23 |
ENSMUST00000208222.2
|
Sesn3
|
sestrin 3 |
| chr11_+_48977888 | 1.18 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr3_+_95434093 | 1.18 |
ENSMUST00000015667.9
ENSMUST00000116304.3 |
Ctss
|
cathepsin S |
| chr3_-_148696155 | 1.15 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr11_+_78234300 | 1.04 |
ENSMUST00000002127.14
ENSMUST00000108295.8 |
Unc119
|
unc-119 lipid binding chaperone |
| chr9_-_116004386 | 1.04 |
ENSMUST00000035014.8
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr1_+_52158599 | 1.02 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr1_-_174749379 | 0.99 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr11_-_101062111 | 0.96 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr11_+_43365103 | 0.96 |
ENSMUST00000173002.8
ENSMUST00000057679.10 |
C1qtnf2
|
C1q and tumor necrosis factor related protein 2 |
| chr1_+_52158693 | 0.94 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr16_-_35759461 | 0.91 |
ENSMUST00000081933.14
ENSMUST00000114885.3 |
Dtx3l
|
deltex 3-like, E3 ubiquitin ligase |
| chr14_+_34097474 | 0.90 |
ENSMUST00000227130.2
|
Mmrn2
|
multimerin 2 |
| chr2_+_71283620 | 0.89 |
ENSMUST00000037210.9
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr5_-_92496730 | 0.85 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr6_-_49191891 | 0.83 |
ENSMUST00000031838.9
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr8_-_85500010 | 0.83 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr1_+_52158721 | 0.82 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr6_-_87312743 | 0.78 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr7_-_18757461 | 0.78 |
ENSMUST00000036018.6
|
Foxa3
|
forkhead box A3 |
| chrX_+_138464065 | 0.77 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr1_+_175459559 | 0.75 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr10_+_21758083 | 0.74 |
ENSMUST00000120509.8
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_35780977 | 0.74 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr1_+_175459735 | 0.71 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr10_+_69048464 | 0.71 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr1_+_16758629 | 0.71 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr8_-_85500998 | 0.70 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr4_-_136613498 | 0.69 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr3_+_90444613 | 0.66 |
ENSMUST00000107335.2
|
S100a16
|
S100 calcium binding protein A16 |
| chr2_-_127630769 | 0.65 |
ENSMUST00000028857.14
ENSMUST00000110357.2 |
Nphp1
|
nephronophthisis 1 (juvenile) homolog (human) |
| chr6_-_39095144 | 0.64 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr14_-_61283911 | 0.64 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_22958956 | 0.59 |
ENSMUST00000226571.2
ENSMUST00000114529.10 ENSMUST00000114526.9 ENSMUST00000228050.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr11_-_77784922 | 0.59 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr11_+_87457479 | 0.59 |
ENSMUST00000239011.2
|
Septin4
|
septin 4 |
| chr7_+_126895463 | 0.59 |
ENSMUST00000106306.9
ENSMUST00000120857.8 |
Itgal
|
integrin alpha L |
| chr16_+_35759346 | 0.58 |
ENSMUST00000023622.13
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr10_+_128542120 | 0.58 |
ENSMUST00000054125.9
|
Pmel
|
premelanosome protein |
| chrX_-_43879055 | 0.57 |
ENSMUST00000060481.9
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr5_-_92653377 | 0.57 |
ENSMUST00000031377.9
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr3_+_90444537 | 0.57 |
ENSMUST00000098911.10
|
S100a16
|
S100 calcium binding protein A16 |
| chr1_+_182236728 | 0.57 |
ENSMUST00000117245.2
|
Trp53bp2
|
transformation related protein 53 binding protein 2 |
| chr3_+_142202642 | 0.56 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr11_-_107361525 | 0.56 |
ENSMUST00000103064.10
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chrX_-_43879022 | 0.54 |
ENSMUST00000115056.2
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr3_-_142101418 | 0.54 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr19_+_56385531 | 0.52 |
ENSMUST00000026062.10
|
Casp7
|
caspase 7 |
| chr10_+_69048506 | 0.51 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr13_-_19579898 | 0.51 |
ENSMUST00000197565.3
ENSMUST00000221380.2 ENSMUST00000200323.3 ENSMUST00000199924.2 ENSMUST00000222869.2 |
Stard3nl
|
STARD3 N-terminal like |
| chr2_+_127967951 | 0.50 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr8_-_106665060 | 0.48 |
ENSMUST00000034369.10
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10 |
| chr7_+_126895423 | 0.47 |
ENSMUST00000117762.8
|
Itgal
|
integrin alpha L |
| chrM_+_9459 | 0.46 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr11_-_3489228 | 0.46 |
ENSMUST00000075118.10
ENSMUST00000136243.2 ENSMUST00000020721.15 ENSMUST00000170588.8 |
Smtn
|
smoothelin |
| chr1_-_155912216 | 0.45 |
ENSMUST00000027738.14
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr17_+_23898223 | 0.45 |
ENSMUST00000024699.4
ENSMUST00000232719.2 |
Cldn6
|
claudin 6 |
| chr13_-_100753419 | 0.45 |
ENSMUST00000168772.2
ENSMUST00000163163.9 ENSMUST00000022137.14 |
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr14_-_73622638 | 0.44 |
ENSMUST00000228637.2
ENSMUST00000022704.9 |
Itm2b
|
integral membrane protein 2B |
| chr5_+_102629240 | 0.44 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr19_+_34560922 | 0.42 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr1_-_155912159 | 0.42 |
ENSMUST00000097527.10
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr13_-_19579961 | 0.41 |
ENSMUST00000039694.13
|
Stard3nl
|
STARD3 N-terminal like |
| chr8_+_105897300 | 0.41 |
ENSMUST00000052209.9
ENSMUST00000109392.9 ENSMUST00000109395.8 |
Cbfb
|
core binding factor beta |
| chr7_+_126895531 | 0.41 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr7_-_89176294 | 0.39 |
ENSMUST00000207932.2
|
Prss23
|
protease, serine 23 |
| chr19_+_41017714 | 0.38 |
ENSMUST00000051806.12
ENSMUST00000112200.3 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr19_+_46611826 | 0.37 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr9_+_108394761 | 0.37 |
ENSMUST00000192819.2
|
Qrich1
|
glutamine-rich 1 |
| chr2_+_22959223 | 0.37 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr13_+_38009981 | 0.36 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr5_-_99391073 | 0.36 |
ENSMUST00000166484.2
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr13_-_100753181 | 0.35 |
ENSMUST00000225754.2
|
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr1_+_61677977 | 0.35 |
ENSMUST00000075374.10
|
Pard3b
|
par-3 family cell polarity regulator beta |
| chr3_-_57202301 | 0.34 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr7_-_4448631 | 0.34 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chrX_+_20714782 | 0.33 |
ENSMUST00000001155.11
ENSMUST00000122312.8 ENSMUST00000120356.8 ENSMUST00000122850.2 |
Araf
|
Araf proto-oncogene, serine/threonine kinase |
| chr13_-_23894697 | 0.33 |
ENSMUST00000091707.13
ENSMUST00000006787.8 |
Hfe
|
homeostatic iron regulator |
| chr19_-_57185988 | 0.31 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr13_-_23894828 | 0.31 |
ENSMUST00000091706.14
|
Hfe
|
homeostatic iron regulator |
| chr1_-_156766957 | 0.30 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr1_+_44158111 | 0.29 |
ENSMUST00000155917.8
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chrM_+_8603 | 0.29 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr17_+_34783242 | 0.28 |
ENSMUST00000015612.14
|
Notch4
|
notch 4 |
| chr19_-_24202344 | 0.27 |
ENSMUST00000099558.5
ENSMUST00000232956.2 |
Tjp2
|
tight junction protein 2 |
| chr11_-_43727071 | 0.26 |
ENSMUST00000167574.2
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr17_+_80597632 | 0.25 |
ENSMUST00000227729.2
ENSMUST00000061703.10 |
Morn2
|
MORN repeat containing 2 |
| chr13_-_113182891 | 0.25 |
ENSMUST00000231962.2
ENSMUST00000022282.6 |
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr7_-_5128936 | 0.25 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr8_+_46338498 | 0.24 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr3_-_102871440 | 0.23 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr18_+_7869705 | 0.22 |
ENSMUST00000166062.8
ENSMUST00000169010.8 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chrX_-_9335525 | 0.22 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr14_+_55815580 | 0.21 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr19_-_57185928 | 0.21 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr1_-_184464810 | 0.21 |
ENSMUST00000048572.7
|
Hlx
|
H2.0-like homeobox |
| chr1_+_16758731 | 0.21 |
ENSMUST00000190366.2
|
Ly96
|
lymphocyte antigen 96 |
| chr2_+_22959452 | 0.21 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr9_-_107419309 | 0.20 |
ENSMUST00000195235.6
|
Cyb561d2
|
cytochrome b-561 domain containing 2 |
| chr19_-_3955230 | 0.20 |
ENSMUST00000145791.8
|
Tcirg1
|
T cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 |
| chr10_-_53952686 | 0.18 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr4_-_129132963 | 0.17 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr14_+_65504067 | 0.16 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr4_-_107035479 | 0.16 |
ENSMUST00000058585.14
|
Tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr14_+_55815999 | 0.14 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr10_-_12424623 | 0.14 |
ENSMUST00000219003.2
|
Utrn
|
utrophin |
| chr10_+_56255184 | 0.14 |
ENSMUST00000220069.2
|
Gja1
|
gap junction protein, alpha 1 |
| chr1_+_57884693 | 0.14 |
ENSMUST00000169772.3
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr18_+_37840092 | 0.14 |
ENSMUST00000195823.2
|
Pcdhga6
|
protocadherin gamma subfamily A, 6 |
| chr14_+_55815879 | 0.14 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr10_+_18283405 | 0.13 |
ENSMUST00000037341.14
|
Nhsl1
|
NHS-like 1 |
| chr2_+_38401826 | 0.13 |
ENSMUST00000112895.8
|
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr6_-_70313491 | 0.13 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr19_-_57185808 | 0.12 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr3_+_89328926 | 0.12 |
ENSMUST00000094378.10
ENSMUST00000137793.2 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr6_-_87312681 | 0.12 |
ENSMUST00000204805.3
|
Antxr1
|
anthrax toxin receptor 1 |
| chr14_+_55815817 | 0.11 |
ENSMUST00000174259.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr11_+_119283887 | 0.11 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr13_-_30170031 | 0.11 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr10_+_94411119 | 0.10 |
ENSMUST00000121471.8
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr5_+_102629365 | 0.10 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr5_+_24598633 | 0.10 |
ENSMUST00000138168.3
ENSMUST00000115077.8 |
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr8_-_46577183 | 0.09 |
ENSMUST00000170416.8
|
Snx25
|
sorting nexin 25 |
| chr13_-_100689212 | 0.09 |
ENSMUST00000022140.12
|
Ocln
|
occludin |
| chr2_+_112114906 | 0.08 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr9_-_35179042 | 0.08 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chrX_+_93679671 | 0.07 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr10_+_82696135 | 0.07 |
ENSMUST00000219442.3
|
Txnrd1
|
thioredoxin reductase 1 |
| chr15_+_102391614 | 0.06 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chrX_+_135608725 | 0.06 |
ENSMUST00000055104.6
|
Tceal1
|
transcription elongation factor A (SII)-like 1 |
| chr1_-_163141278 | 0.06 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr11_+_48977495 | 0.06 |
ENSMUST00000152914.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr19_+_34585322 | 0.06 |
ENSMUST00000076249.6
|
Ifit3b
|
interferon-induced protein with tetratricopeptide repeats 3B |
| chr12_-_113542610 | 0.05 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr1_+_57445487 | 0.04 |
ENSMUST00000027114.6
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr12_+_108601963 | 0.04 |
ENSMUST00000223109.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr16_-_43484494 | 0.04 |
ENSMUST00000096065.6
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr15_-_66432938 | 0.04 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr2_+_38401655 | 0.04 |
ENSMUST00000054234.10
ENSMUST00000112902.8 |
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr11_+_87938626 | 0.04 |
ENSMUST00000107920.10
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr3_+_152052102 | 0.03 |
ENSMUST00000117492.9
ENSMUST00000026507.13 ENSMUST00000197748.5 |
Usp33
|
ubiquitin specific peptidase 33 |
| chr8_+_46338557 | 0.02 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr14_-_78970160 | 0.02 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr7_-_140846328 | 0.02 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chrX_-_100266032 | 0.00 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr14_+_65504151 | 0.00 |
ENSMUST00000169656.3
ENSMUST00000226005.2 |
Fbxo16
|
F-box protein 16 |
| chr18_+_37637317 | 0.00 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.8 | 3.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.6 | 2.4 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.4 | 1.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.3 | 1.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.3 | 1.5 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.3 | 1.0 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 1.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.7 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 0.9 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 0.6 | GO:1900390 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.2 | 1.7 | GO:0031179 | peptide modification(GO:0031179) |
| 0.2 | 0.6 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.2 | 0.8 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.2 | 3.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 2.4 | GO:0070863 | positive regulation of protein glycosylation(GO:0060050) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 1.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.9 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.9 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 2.4 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 1.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.5 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 1.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.3 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.1 | 1.8 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 1.5 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.6 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 1.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.9 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 2.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.4 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 1.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.6 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 1.0 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.3 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.6 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.9 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 5.8 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.0 | 0.6 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 1.9 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.5 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.2 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.7 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.2 | 2.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.8 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 3.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 6.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.4 | 1.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 1.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.3 | 1.5 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 2.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 3.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 2.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 0.9 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.0 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.6 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 3.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 6.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.8 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 3.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 4.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 6.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 3.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 0.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 2.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 6.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |