Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
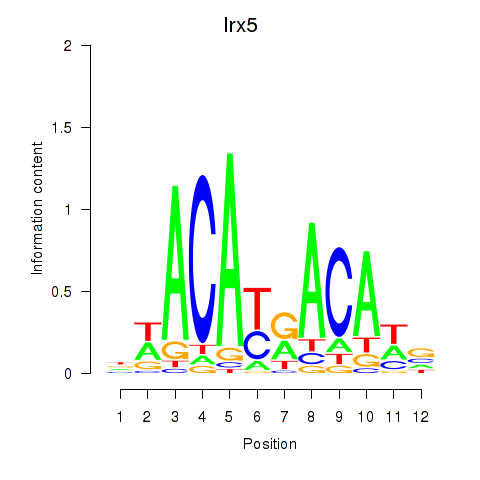
Results for Irx5
Z-value: 1.31
Transcription factors associated with Irx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx5
|
ENSMUSG00000031737.12 | Iroquois homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx5 | mm39_v1_chr8_+_93084253_93084379 | -0.24 | 1.6e-01 | Click! |
Activity profile of Irx5 motif
Sorted Z-values of Irx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87074380 | 19.29 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr5_-_87485023 | 19.00 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr15_+_82336535 | 13.00 |
ENSMUST00000089129.7
ENSMUST00000229313.2 ENSMUST00000231136.2 |
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr7_-_14172434 | 12.44 |
ENSMUST00000210396.2
ENSMUST00000168252.9 |
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr15_-_82648376 | 12.39 |
ENSMUST00000055721.6
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr2_+_172994841 | 11.78 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr5_-_87402659 | 10.45 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr19_-_7779943 | 7.91 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr6_+_71470987 | 6.68 |
ENSMUST00000114179.3
|
Rnf103
|
ring finger protein 103 |
| chr3_+_20011201 | 6.66 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chrX_+_20416019 | 6.24 |
ENSMUST00000023832.7
|
Rgn
|
regucalcin |
| chr3_+_20011251 | 5.20 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr3_+_20011405 | 4.59 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr15_-_82278223 | 4.33 |
ENSMUST00000170255.2
|
Cyp2d11
|
cytochrome P450, family 2, subfamily d, polypeptide 11 |
| chr4_+_104623505 | 4.21 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr18_+_56533389 | 3.72 |
ENSMUST00000237355.2
ENSMUST00000237422.2 |
Gramd3
|
GRAM domain containing 3 |
| chr3_+_63203235 | 3.10 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr13_+_25127127 | 3.07 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr2_+_103858066 | 2.78 |
ENSMUST00000028603.10
|
Fbxo3
|
F-box protein 3 |
| chr7_-_133384449 | 2.63 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr4_+_141473983 | 2.58 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr10_+_86614864 | 2.40 |
ENSMUST00000099396.3
|
Nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr8_-_73188887 | 2.36 |
ENSMUST00000109974.2
|
Calr3
|
calreticulin 3 |
| chr19_+_39499288 | 2.29 |
ENSMUST00000025968.5
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr11_-_110142565 | 2.25 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr14_+_48358267 | 2.19 |
ENSMUST00000073150.6
|
Peli2
|
pellino 2 |
| chr4_+_8535604 | 2.16 |
ENSMUST00000060232.8
|
Rab2a
|
RAB2A, member RAS oncogene family |
| chr4_-_131871797 | 2.05 |
ENSMUST00000056336.2
|
Oprd1
|
opioid receptor, delta 1 |
| chr2_-_155661324 | 2.01 |
ENSMUST00000124586.2
|
BC029722
|
cDNA sequence BC029722 |
| chr2_+_103858168 | 2.01 |
ENSMUST00000111135.8
ENSMUST00000111136.8 ENSMUST00000102565.4 |
Fbxo3
|
F-box protein 3 |
| chr7_+_102289455 | 1.92 |
ENSMUST00000098221.2
|
Olfr554
|
olfactory receptor 554 |
| chr2_-_101451383 | 1.87 |
ENSMUST00000090513.11
|
Iftap
|
intraflagellar transport associated protein |
| chr11_-_69286159 | 1.83 |
ENSMUST00000108660.8
ENSMUST00000051620.5 |
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr12_+_85335365 | 1.81 |
ENSMUST00000059341.5
|
Zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr6_+_50087826 | 1.54 |
ENSMUST00000167628.2
|
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr5_+_107655487 | 1.52 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr11_-_46280298 | 1.41 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chrX_+_37861548 | 1.36 |
ENSMUST00000050744.6
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr11_-_46280281 | 1.30 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr1_-_169796709 | 1.27 |
ENSMUST00000027989.13
ENSMUST00000111353.4 |
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr11_+_69286473 | 1.23 |
ENSMUST00000144531.2
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr16_-_30152708 | 1.17 |
ENSMUST00000229503.2
|
Atp13a3
|
ATPase type 13A3 |
| chr3_+_138911648 | 1.08 |
ENSMUST00000062306.7
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr4_-_114991174 | 1.03 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr11_+_78067537 | 1.03 |
ENSMUST00000098545.12
|
Tlcd1
|
TLC domain containing 1 |
| chr11_-_121410152 | 1.02 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr12_-_113223839 | 0.98 |
ENSMUST00000194738.6
ENSMUST00000178282.3 |
Igha
|
immunoglobulin heavy constant alpha |
| chr18_-_35348049 | 0.90 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr14_+_48683797 | 0.89 |
ENSMUST00000111735.10
|
Tmem260
|
transmembrane protein 260 |
| chr1_+_86354045 | 0.87 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr9_+_32305259 | 0.83 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr14_-_4506874 | 0.80 |
ENSMUST00000224934.2
|
Thrb
|
thyroid hormone receptor beta |
| chr8_-_58249608 | 0.80 |
ENSMUST00000204067.3
|
Galntl6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr12_-_85335193 | 0.71 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr3_-_88669551 | 0.70 |
ENSMUST00000183267.2
|
Syt11
|
synaptotagmin XI |
| chr19_-_28945194 | 0.67 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like |
| chr4_+_111830119 | 0.65 |
ENSMUST00000106568.8
ENSMUST00000055014.11 ENSMUST00000163281.2 |
Skint7
|
selection and upkeep of intraepithelial T cells 7 |
| chr17_+_72076728 | 0.59 |
ENSMUST00000230305.2
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr11_+_42312150 | 0.58 |
ENSMUST00000192403.2
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr6_+_56974228 | 0.57 |
ENSMUST00000228285.2
ENSMUST00000227847.2 ENSMUST00000226689.2 ENSMUST00000227131.2 ENSMUST00000227631.2 ENSMUST00000227188.2 |
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr16_+_3408906 | 0.53 |
ENSMUST00000216259.2
|
Olfr161
|
olfactory receptor 161 |
| chr9_+_110948492 | 0.53 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr19_+_26600820 | 0.52 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_-_35954573 | 0.49 |
ENSMUST00000095467.4
|
Mucl3
|
mucin like 3 |
| chr7_+_55889488 | 0.43 |
ENSMUST00000032633.12
ENSMUST00000155533.2 ENSMUST00000156886.8 |
Oca2
|
oculocutaneous albinism II |
| chr17_+_72076678 | 0.43 |
ENSMUST00000230427.2
ENSMUST00000229952.2 ENSMUST00000230333.2 |
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr18_-_64794338 | 0.41 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr11_+_73244561 | 0.40 |
ENSMUST00000108465.4
|
Olfr20
|
olfactory receptor 20 |
| chr5_-_46013838 | 0.36 |
ENSMUST00000087164.10
ENSMUST00000121573.8 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr3_-_32419609 | 0.36 |
ENSMUST00000139660.2
ENSMUST00000168566.3 ENSMUST00000029199.11 |
Zmat3
|
zinc finger matrin type 3 |
| chr15_-_81742686 | 0.35 |
ENSMUST00000050467.9
ENSMUST00000231000.2 |
Tob2
|
transducer of ERBB2, 2 |
| chr14_+_54380308 | 0.34 |
ENSMUST00000196323.2
|
Trdc
|
T cell receptor delta, constant region |
| chr4_-_89229256 | 0.34 |
ENSMUST00000097981.6
|
Cdkn2b
|
cyclin dependent kinase inhibitor 2B |
| chr2_-_136229849 | 0.27 |
ENSMUST00000035264.9
ENSMUST00000077200.4 |
Pak5
|
p21 (RAC1) activated kinase 5 |
| chr17_+_87332978 | 0.27 |
ENSMUST00000024959.10
|
Cript
|
cysteine-rich PDZ-binding protein |
| chr13_+_27496593 | 0.26 |
ENSMUST00000091680.9
|
Prl6a1
|
prolactin family 6, subfamily a, member 1 |
| chr6_-_113694633 | 0.22 |
ENSMUST00000204533.3
|
Ghrl
|
ghrelin |
| chr4_-_43700807 | 0.19 |
ENSMUST00000055545.5
|
Olfr70
|
olfactory receptor 70 |
| chr11_-_99176086 | 0.19 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24 |
| chr6_-_129303659 | 0.17 |
ENSMUST00000203159.2
|
Clec2m
|
C-type lectin domain family 2, member m |
| chr6_-_57938488 | 0.16 |
ENSMUST00000228097.2
|
Vmn1r24
|
vomeronasal 1 receptor 24 |
| chr6_+_56926317 | 0.11 |
ENSMUST00000227073.2
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr2_+_152269532 | 0.09 |
ENSMUST00000128737.2
|
6820408C15Rik
|
RIKEN cDNA 6820408C15 gene |
| chr1_-_132635078 | 0.06 |
ENSMUST00000187861.7
|
Nfasc
|
neurofascin |
| chr18_+_35347983 | 0.06 |
ENSMUST00000235449.2
ENSMUST00000235269.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr16_-_22630327 | 0.06 |
ENSMUST00000040592.6
|
Crygs
|
crystallin, gamma S |
| chr7_-_86016045 | 0.06 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr10_-_43934774 | 0.04 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr4_+_112089442 | 0.03 |
ENSMUST00000038455.12
ENSMUST00000170945.2 |
Skint3
|
selection and upkeep of intraepithelial T cells 3 |
| chr4_+_112470794 | 0.03 |
ENSMUST00000058791.14
ENSMUST00000186969.2 |
Skint2
|
selection and upkeep of intraepithelial T cells 2 |
| chr6_-_57306479 | 0.02 |
ENSMUST00000227283.2
ENSMUST00000228356.2 |
Vmn1r16
|
vomeronasal 1 receptor 16 |
| chr14_+_8348779 | 0.00 |
ENSMUST00000022256.5
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.3 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 2.9 | 11.8 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.6 | 6.2 | GO:1903630 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 1.4 | 19.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.8 | 3.1 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.5 | 7.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 16.5 | GO:0046688 | copper ion transport(GO:0006825) response to copper ion(GO:0046688) |
| 0.4 | 3.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 2.6 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.4 | 30.7 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.3 | 2.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 4.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 2.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 0.7 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 0.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 2.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.5 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.2 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.1 | 0.5 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 6.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.4 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 1.0 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.3 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 1.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 2.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 2.4 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 1.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 16.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 3.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 3.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 9.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.5 | 16.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.2 | 48.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 20.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.5 | 7.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 3.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 2.0 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 11.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 2.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 0.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 1.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 2.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 2.6 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 2.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 16.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 11.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 16.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 2.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 4.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |