Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Irx6_Irx2_Irx3
Z-value: 0.76
Transcription factors associated with Irx6_Irx2_Irx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
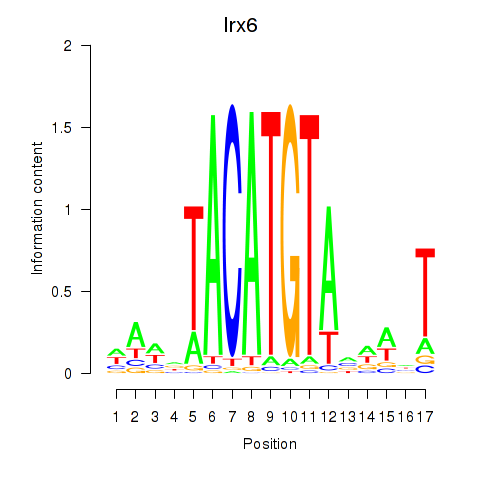
Irx6
|
ENSMUSG00000031738.15 | Iroquois homeobox 6 |
|
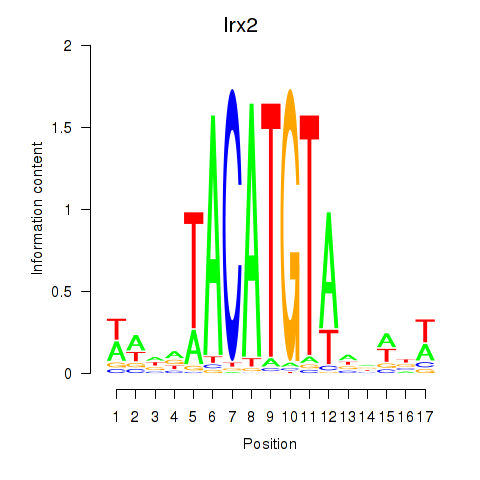
Irx2
|
ENSMUSG00000001504.11 | Iroquois homeobox 2 |
|
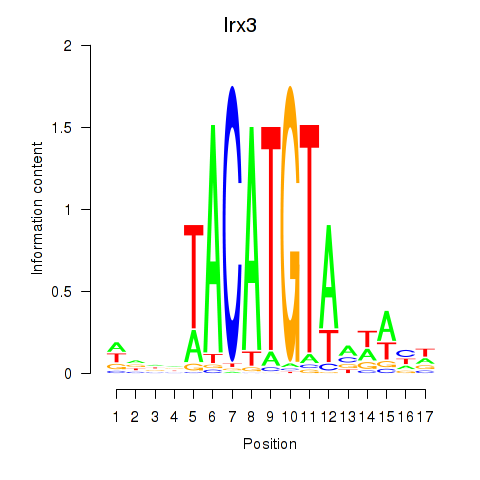
Irx3
|
ENSMUSG00000031734.14 | Iroquois related homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx3 | mm39_v1_chr8_-_92528543_92528569 | 0.04 | 8.2e-01 | Click! |
| Irx6 | mm39_v1_chr8_+_93400916_93400916 | -0.03 | 8.7e-01 | Click! |
| Irx2 | mm39_v1_chr13_+_72776921_72776967 | -0.01 | 9.3e-01 | Click! |
Activity profile of Irx6_Irx2_Irx3 motif
Sorted Z-values of Irx6_Irx2_Irx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_16964801 | 2.27 |
ENSMUST00000232479.2
ENSMUST00000232344.2 ENSMUST00000069064.7 |
Ydjc
|
YdjC homolog (bacterial) |
| chr19_+_20579322 | 1.49 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr4_+_63262775 | 1.45 |
ENSMUST00000030044.3
|
Orm1
|
orosomucoid 1 |
| chr4_-_62005498 | 1.36 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr4_-_138858340 | 1.36 |
ENSMUST00000143971.2
|
Micos10
|
mitochondrial contact site and cristae organizing system subunit 10 |
| chr16_+_49620883 | 1.31 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr4_-_60455331 | 1.29 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr16_-_37681508 | 1.26 |
ENSMUST00000205931.2
|
Gm36028
|
predicted gene, 36028 |
| chr5_-_87402659 | 1.24 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr8_+_86219191 | 1.17 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr8_-_79539838 | 1.09 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr18_-_36903237 | 1.08 |
ENSMUST00000235494.2
|
Hars
|
histidyl-tRNA synthetase |
| chr17_+_46807637 | 1.02 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr6_-_56878854 | 0.99 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr6_-_130363837 | 0.90 |
ENSMUST00000032288.6
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr4_+_97665843 | 0.88 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr17_-_27352876 | 0.82 |
ENSMUST00000119227.3
ENSMUST00000025045.15 |
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr15_-_35938155 | 0.80 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr9_-_48516447 | 0.78 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr11_+_100211363 | 0.78 |
ENSMUST00000152521.2
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr7_-_103492361 | 0.77 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr13_-_22219738 | 0.76 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr8_-_86091970 | 0.74 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr15_-_35938328 | 0.73 |
ENSMUST00000014457.15
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr17_-_30845845 | 0.73 |
ENSMUST00000235547.2
ENSMUST00000188852.2 |
Glo1
1700097N02Rik
|
glyoxalase 1 RIKEN cDNA 1700097N02 gene |
| chr18_+_36797113 | 0.72 |
ENSMUST00000036765.8
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr13_-_21937997 | 0.69 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15 |
| chr14_-_99231754 | 0.68 |
ENSMUST00000081987.5
|
Rpl36a-ps1
|
ribosomal protein L36A, pseudogene 1 |
| chr9_-_19452372 | 0.66 |
ENSMUST00000213834.2
|
Olfr853
|
olfactory receptor 853 |
| chr6_+_121983720 | 0.64 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chr4_+_154226819 | 0.60 |
ENSMUST00000030895.12
|
Wrap73
|
WD repeat containing, antisense to Trp73 |
| chr9_+_38684364 | 0.56 |
ENSMUST00000217114.3
ENSMUST00000213958.3 |
Olfr921
|
olfactory receptor 921 |
| chr6_-_123627684 | 0.56 |
ENSMUST00000170808.3
|
Vmn2r22
|
vomeronasal 2, receptor 22 |
| chr19_-_39451509 | 0.55 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr6_-_116561156 | 0.55 |
ENSMUST00000061723.6
|
Olfr215
|
olfactory receptor 215 |
| chr6_-_83294526 | 0.52 |
ENSMUST00000005810.9
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr1_-_181985663 | 0.51 |
ENSMUST00000169123.4
|
Vmn1r1
|
vomeronasal 1 receptor 1 |
| chr14_-_9015639 | 0.51 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr7_-_130148984 | 0.50 |
ENSMUST00000160289.9
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr13_+_73479101 | 0.50 |
ENSMUST00000022098.10
ENSMUST00000222030.2 |
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr17_+_23819818 | 0.50 |
ENSMUST00000095595.9
ENSMUST00000115509.8 ENSMUST00000120967.8 ENSMUST00000148062.8 ENSMUST00000129227.8 |
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr7_+_106413336 | 0.49 |
ENSMUST00000166880.3
ENSMUST00000075414.8 |
Olfr701
|
olfactory receptor 701 |
| chr10_-_23663109 | 0.49 |
ENSMUST00000218221.2
ENSMUST00000218107.2 |
Rps12
|
ribosomal protein S12 |
| chr9_+_64080644 | 0.48 |
ENSMUST00000034966.9
|
Rpl4
|
ribosomal protein L4 |
| chr2_+_130975417 | 0.47 |
ENSMUST00000110225.2
|
Gm11037
|
predicted gene 11037 |
| chr17_+_23819861 | 0.47 |
ENSMUST00000123866.8
|
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr11_-_102787950 | 0.46 |
ENSMUST00000067444.10
|
Gfap
|
glial fibrillary acidic protein |
| chr13_-_23727549 | 0.45 |
ENSMUST00000224359.2
|
H2bc9
|
H2B clustered histone 9 |
| chr13_+_83720457 | 0.45 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr6_-_85797946 | 0.45 |
ENSMUST00000032074.5
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr8_-_86107593 | 0.45 |
ENSMUST00000122452.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr2_-_89951611 | 0.44 |
ENSMUST00000216493.2
ENSMUST00000214404.2 |
Olfr1269
|
olfactory receptor 1269 |
| chr19_-_7688628 | 0.44 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr5_-_44139099 | 0.43 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr10_+_84412490 | 0.43 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr10_+_12881087 | 0.43 |
ENSMUST00000105139.5
|
Sf3b5
|
splicing factor 3b, subunit 5 |
| chr13_+_83720484 | 0.43 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr4_-_88595161 | 0.42 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16 |
| chr13_+_74554509 | 0.42 |
ENSMUST00000222435.2
|
Ftl1-ps1
|
ferritin light polypeptide 1, pseudogene 1 |
| chr5_-_44139121 | 0.42 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr14_+_54068562 | 0.42 |
ENSMUST00000103673.11
ENSMUST00000186573.2 |
Trav18
|
T cell receptor alpha variable 18 |
| chr2_-_52225146 | 0.42 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr8_+_57004125 | 0.40 |
ENSMUST00000110322.9
ENSMUST00000040218.13 ENSMUST00000210863.2 |
Fbxo8
|
F-box protein 8 |
| chr6_+_57234937 | 0.40 |
ENSMUST00000228297.2
|
Vmn1r15
|
vomeronasal 1 receptor 15 |
| chr13_+_4624074 | 0.39 |
ENSMUST00000021628.4
|
Akr1c21
|
aldo-keto reductase family 1, member C21 |
| chrX_-_142716085 | 0.39 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr7_+_28050077 | 0.39 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr12_-_115825934 | 0.39 |
ENSMUST00000198777.2
|
Ighv1-77
|
immunoglobulin heavy variable 1-77 |
| chr5_-_144944838 | 0.39 |
ENSMUST00000116454.10
|
Kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr1_-_174078542 | 0.38 |
ENSMUST00000061990.5
|
Olfr419
|
olfactory receptor 419 |
| chr5_-_137246852 | 0.38 |
ENSMUST00000179412.2
|
Muc3a
|
mucin 3A, cell surface associated |
| chr3_-_98471301 | 0.38 |
ENSMUST00000058728.10
|
Gm10681
|
predicted gene 10681 |
| chr3_+_27371206 | 0.38 |
ENSMUST00000174840.2
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr13_+_76727787 | 0.37 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr1_-_128256048 | 0.37 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr2_-_98497609 | 0.37 |
ENSMUST00000099683.2
|
Gm10800
|
predicted gene 10800 |
| chr6_+_124001527 | 0.37 |
ENSMUST00000032238.5
|
Vmn2r26
|
vomeronasal 2, receptor 26 |
| chr4_+_119052548 | 0.37 |
ENSMUST00000106345.3
|
Svbp
|
small vasohibin binding protein |
| chr7_-_102566717 | 0.37 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr2_-_111908072 | 0.37 |
ENSMUST00000213577.2
ENSMUST00000216071.2 |
Olfr1313
|
olfactory receptor 1313 |
| chr10_+_61007733 | 0.36 |
ENSMUST00000122261.8
ENSMUST00000121297.8 ENSMUST00000035894.12 |
Tbata
|
thymus, brain and testes associated |
| chr13_-_24945844 | 0.36 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chr5_+_115417725 | 0.36 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr7_-_103477126 | 0.36 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr7_+_12893495 | 0.36 |
ENSMUST00000236472.2
ENSMUST00000237109.2 |
Vmn1r88
|
vomeronasal 1 receptor, 88 |
| chr12_+_119407145 | 0.36 |
ENSMUST00000048880.7
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr14_+_53550361 | 0.36 |
ENSMUST00000179997.2
|
Trav5n-4
|
T cell receptor alpha variable 5N-4 |
| chr2_+_85715984 | 0.36 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr4_+_98614971 | 0.35 |
ENSMUST00000152889.9
ENSMUST00000154279.3 |
L1td1
|
LINE-1 type transposase domain containing 1 |
| chr8_+_85635189 | 0.35 |
ENSMUST00000003910.13
ENSMUST00000109744.8 |
Dnase2a
|
deoxyribonuclease II alpha |
| chr13_+_23728222 | 0.35 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr8_-_25215778 | 0.35 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr15_+_43340609 | 0.35 |
ENSMUST00000022962.8
|
Emc2
|
ER membrane protein complex subunit 2 |
| chr15_+_91722524 | 0.35 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr7_-_3551003 | 0.35 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr1_+_152626273 | 0.35 |
ENSMUST00000068875.5
|
Apobec4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr7_-_102998876 | 0.34 |
ENSMUST00000215042.2
|
Olfr600
|
olfactory receptor 600 |
| chr5_+_94224244 | 0.34 |
ENSMUST00000139102.2
|
Gm6351
|
predicted gene 6351 |
| chr1_-_173948728 | 0.34 |
ENSMUST00000214446.2
|
Olfr231
|
olfactory receptor 231 |
| chr7_+_23274951 | 0.34 |
ENSMUST00000228832.2
ENSMUST00000227547.2 ENSMUST00000226669.2 ENSMUST00000227932.2 |
Vmn1r169
|
vomeronasal 1 receptor 169 |
| chr8_-_25215856 | 0.34 |
ENSMUST00000033958.15
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr7_-_24033346 | 0.34 |
ENSMUST00000069562.6
|
Tescl
|
tescalcin-like |
| chr4_-_59960659 | 0.34 |
ENSMUST00000075973.3
|
Mup4
|
major urinary protein 4 |
| chr6_-_68857658 | 0.34 |
ENSMUST00000198756.2
|
Gm42543
|
predicted gene 42543 |
| chr6_+_28423539 | 0.33 |
ENSMUST00000020717.12
ENSMUST00000169841.2 |
Arf5
|
ADP-ribosylation factor 5 |
| chr9_-_44255456 | 0.32 |
ENSMUST00000077353.15
|
Hmbs
|
hydroxymethylbilane synthase |
| chr17_+_37710293 | 0.32 |
ENSMUST00000216844.2
ENSMUST00000215974.2 ENSMUST00000215894.2 |
Olfr107
|
olfactory receptor 107 |
| chr8_-_26505605 | 0.32 |
ENSMUST00000016138.11
|
Fnta
|
farnesyltransferase, CAAX box, alpha |
| chr13_-_59890564 | 0.32 |
ENSMUST00000055343.3
|
1700014D04Rik
|
RIKEN cDNA 1700014D04 gene |
| chr19_-_12901783 | 0.32 |
ENSMUST00000213713.2
ENSMUST00000216888.2 ENSMUST00000213177.2 |
Olfr1448
|
olfactory receptor 1448 |
| chr9_-_112061517 | 0.32 |
ENSMUST00000035085.12
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr8_+_85598734 | 0.32 |
ENSMUST00000170296.2
ENSMUST00000136026.8 |
Syce2
|
synaptonemal complex central element protein 2 |
| chr2_+_109522781 | 0.32 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr13_-_98152768 | 0.31 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr2_-_111253457 | 0.31 |
ENSMUST00000213210.2
ENSMUST00000184954.4 |
Olfr1286
|
olfactory receptor 1286 |
| chrX_+_143253677 | 0.31 |
ENSMUST00000178233.2
|
Trpc5os
|
transient receptor potential cation channel, subfamily C, member 5, opposite strand |
| chr2_-_120946877 | 0.31 |
ENSMUST00000110675.3
|
Tgm7
|
transglutaminase 7 |
| chr2_+_85780781 | 0.31 |
ENSMUST00000080698.3
|
Olfr1028
|
olfactory receptor 1028 |
| chr6_-_128700022 | 0.31 |
ENSMUST00000112110.4
|
Klrb1
|
killer cell lectin-like receptor subfamily B member 1 |
| chr7_+_23330147 | 0.31 |
ENSMUST00000227774.2
ENSMUST00000226771.2 ENSMUST00000228681.2 ENSMUST00000228559.2 ENSMUST00000228674.2 ENSMUST00000227866.2 ENSMUST00000227386.2 ENSMUST00000228484.2 ENSMUST00000226321.2 ENSMUST00000226128.2 ENSMUST00000226733.2 ENSMUST00000228228.2 |
Vmn1r171
|
vomeronasal 1 receptor 171 |
| chr6_-_70036183 | 0.31 |
ENSMUST00000197429.5
ENSMUST00000103376.3 |
Igkv7-33
|
immunoglobulin kappa chain variable 7-33 |
| chrX_-_5381319 | 0.31 |
ENSMUST00000058404.5
|
Mycs
|
myc-like oncogene, s-myc protein |
| chr16_+_19302444 | 0.31 |
ENSMUST00000216465.2
ENSMUST00000213531.2 |
Olfr166
|
olfactory receptor 166 |
| chr7_+_120442048 | 0.30 |
ENSMUST00000047875.16
|
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr13_-_27766147 | 0.30 |
ENSMUST00000006664.8
ENSMUST00000095926.2 |
Prl8a1
|
prolactin family 8, subfamily a, member 1 |
| chr19_-_6885657 | 0.30 |
ENSMUST00000149261.8
|
Prdx5
|
peroxiredoxin 5 |
| chr1_-_72739691 | 0.30 |
ENSMUST00000211837.2
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr7_-_23536797 | 0.30 |
ENSMUST00000226319.2
ENSMUST00000228793.2 ENSMUST00000228280.2 ENSMUST00000227661.2 ENSMUST00000226767.2 ENSMUST00000227129.2 |
Vmn1r176
|
vomeronasal 1 receptor 176 |
| chr3_+_59832635 | 0.30 |
ENSMUST00000049476.3
|
Aadacl2fm1
|
AADACL2 family member 1 |
| chr4_-_129229159 | 0.30 |
ENSMUST00000102598.4
|
Rbbp4
|
retinoblastoma binding protein 4, chromatin remodeling factor |
| chr6_-_123395075 | 0.30 |
ENSMUST00000172199.3
|
Vmn2r20
|
vomeronasal 2, receptor 20 |
| chrX_+_9751861 | 0.30 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr5_-_6926523 | 0.30 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr6_+_149031668 | 0.29 |
ENSMUST00000087348.4
|
Gm10203
|
predicted gene 10203 |
| chr13_+_19394484 | 0.29 |
ENSMUST00000200495.2
|
Trgj1
|
T cell receptor gamma joining 1 |
| chr8_-_105368298 | 0.29 |
ENSMUST00000093234.5
|
Ciao2b
|
cytosolic iron-sulfur assembly component 2B |
| chr13_+_67927802 | 0.29 |
ENSMUST00000220570.2
ENSMUST00000164936.10 ENSMUST00000181319.2 |
Zfp493
|
zinc finger protein 493 |
| chr13_-_22375311 | 0.29 |
ENSMUST00000238109.2
|
Vmn1r192
|
vomeronasal 1 receptor 192 |
| chr11_+_73158214 | 0.29 |
ENSMUST00000049676.3
|
Trpv3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr16_-_45313244 | 0.29 |
ENSMUST00000232138.2
|
Gm609
|
predicted gene 609 |
| chr17_+_37710117 | 0.29 |
ENSMUST00000215947.2
|
Olfr107
|
olfactory receptor 107 |
| chr3_+_89366632 | 0.29 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase |
| chr9_+_35819708 | 0.28 |
ENSMUST00000176049.2
ENSMUST00000176153.2 |
Pate13
|
prostate and testis expressed 13 |
| chr1_+_151068127 | 0.28 |
ENSMUST00000224330.2
|
Gm8947
|
predicted gene 8947 |
| chr18_-_78100610 | 0.28 |
ENSMUST00000170760.3
|
Siglec15
|
sialic acid binding Ig-like lectin 15 |
| chr2_+_36575800 | 0.28 |
ENSMUST00000213258.2
|
Olfr346
|
olfactory receptor 346 |
| chr12_-_74363168 | 0.28 |
ENSMUST00000110441.2
|
Gm11042
|
predicted gene 11042 |
| chrX_-_142716200 | 0.28 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr1_-_72739704 | 0.28 |
ENSMUST00000053499.6
ENSMUST00000212710.2 ENSMUST00000212573.2 |
Ankar
|
ankyrin and armadillo repeat containing |
| chr6_-_124410452 | 0.28 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr11_-_53508160 | 0.28 |
ENSMUST00000150568.8
|
Il4
|
interleukin 4 |
| chrX_+_106132055 | 0.28 |
ENSMUST00000150494.2
|
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr1_+_161322219 | 0.27 |
ENSMUST00000086084.2
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr6_+_123679629 | 0.27 |
ENSMUST00000172391.4
|
Vmn2r23
|
vomeronasal 2, receptor 23 |
| chr9_-_108305683 | 0.27 |
ENSMUST00000076592.4
|
Iho1
|
interactor of HORMAD1 1 |
| chr5_-_87847268 | 0.27 |
ENSMUST00000196869.5
ENSMUST00000199624.5 ENSMUST00000198057.5 ENSMUST00000082370.10 |
Csn2
|
casein beta |
| chr13_-_27852017 | 0.27 |
ENSMUST00000006660.7
|
Prl7a2
|
prolactin family 7, subfamily a, member 2 |
| chr5_+_123214332 | 0.27 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr11_-_82719850 | 0.27 |
ENSMUST00000021036.13
ENSMUST00000074515.11 ENSMUST00000103218.3 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr11_-_118021460 | 0.27 |
ENSMUST00000132685.9
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr19_+_12647803 | 0.27 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr11_-_72106418 | 0.27 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr12_-_87691390 | 0.27 |
ENSMUST00000220528.2
|
Ubl5b
|
ubiquitin-like 5B |
| chr6_-_57821483 | 0.26 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr12_-_81468705 | 0.26 |
ENSMUST00000085319.4
|
Adam4
|
a disintegrin and metallopeptidase domain 4 |
| chr11_+_11414256 | 0.26 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr13_+_22534534 | 0.26 |
ENSMUST00000226909.2
ENSMUST00000227167.2 ENSMUST00000226786.2 |
Vmn1r198
|
vomeronasal 1 receptor 198 |
| chr6_+_52690714 | 0.26 |
ENSMUST00000080723.11
ENSMUST00000149588.8 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr6_+_89860642 | 0.26 |
ENSMUST00000226345.2
ENSMUST00000204656.3 ENSMUST00000227456.2 ENSMUST00000227047.2 ENSMUST00000226120.2 ENSMUST00000227888.2 ENSMUST00000228183.2 ENSMUST00000227625.2 |
Vmn1r44
|
vomeronasal 1 receptor 44 |
| chr5_-_137599304 | 0.26 |
ENSMUST00000037620.14
ENSMUST00000154708.8 |
Mospd3
|
motile sperm domain containing 3 |
| chr17_+_30845909 | 0.26 |
ENSMUST00000236140.2
ENSMUST00000236118.2 ENSMUST00000235390.2 |
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr8_+_111782951 | 0.26 |
ENSMUST00000210390.2
|
Exosc6
|
exosome component 6 |
| chr14_+_53836282 | 0.26 |
ENSMUST00000103655.3
|
Trav4-3
|
T cell receptor alpha variable 4-3 |
| chr13_+_91071077 | 0.26 |
ENSMUST00000051955.9
|
Rps23
|
ribosomal protein S23 |
| chr7_+_5221492 | 0.26 |
ENSMUST00000228062.2
ENSMUST00000227798.2 |
Vmn1r57
|
vomeronasal 1 receptor 57 |
| chr14_+_53859114 | 0.25 |
ENSMUST00000103657.6
|
Trav12-3
|
T cell receptor alpha variable 12-3 |
| chr10_-_129565161 | 0.25 |
ENSMUST00000204717.3
ENSMUST00000216794.2 ENSMUST00000217219.2 |
Olfr805
|
olfactory receptor 805 |
| chr16_-_45313324 | 0.25 |
ENSMUST00000114585.3
|
Gm609
|
predicted gene 609 |
| chr9_+_37765565 | 0.25 |
ENSMUST00000213956.2
|
Olfr877
|
olfactory receptor 877 |
| chr11_-_102787972 | 0.25 |
ENSMUST00000077902.5
|
Gfap
|
glial fibrillary acidic protein |
| chr14_-_68771138 | 0.25 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr2_+_36308959 | 0.25 |
ENSMUST00000216645.2
|
Olfr339
|
olfactory receptor 339 |
| chr7_-_23206631 | 0.25 |
ENSMUST00000227713.2
|
Vmn1r167
|
vomeronasal 1 receptor 167 |
| chr3_-_57559088 | 0.25 |
ENSMUST00000160959.8
|
Commd2
|
COMM domain containing 2 |
| chr19_-_32443978 | 0.25 |
ENSMUST00000078034.5
|
Rpl9-ps6
|
ribosomal protein L9, pseudogene 6 |
| chr3_-_151960992 | 0.25 |
ENSMUST00000198750.5
|
Nexn
|
nexilin |
| chr11_-_87716849 | 0.25 |
ENSMUST00000103177.10
|
Lpo
|
lactoperoxidase |
| chr9_-_35481689 | 0.24 |
ENSMUST00000115110.5
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr2_+_86655007 | 0.24 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr1_+_75377616 | 0.24 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr14_+_53478202 | 0.24 |
ENSMUST00000179583.3
|
Trav12n-3
|
T cell receptor alpha variable 12N-3 |
| chr7_-_12853779 | 0.24 |
ENSMUST00000227220.2
ENSMUST00000227700.2 ENSMUST00000226604.2 |
Vmn1r86
|
vomeronasal 1 receptor 86 |
| chr8_-_86091946 | 0.24 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr7_+_6441687 | 0.24 |
ENSMUST00000218906.2
|
Olfr1344
|
olfactory receptor 1344 |
| chr6_-_130208601 | 0.24 |
ENSMUST00000088011.11
ENSMUST00000112013.8 ENSMUST00000049304.14 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr4_-_49506538 | 0.24 |
ENSMUST00000043056.9
|
Baat
|
bile acid-Coenzyme A: amino acid N-acyltransferase |
| chr1_-_38875757 | 0.24 |
ENSMUST00000147695.9
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr10_-_90959853 | 0.24 |
ENSMUST00000170810.8
ENSMUST00000076694.13 |
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr17_-_40630096 | 0.23 |
ENSMUST00000026498.5
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chrX_-_48823936 | 0.23 |
ENSMUST00000215373.3
|
Olfr1321
|
olfactory receptor 1321 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx6_Irx2_Irx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.3 | 1.2 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.2 | 1.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.4 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 1.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.3 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 1.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.3 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 1.0 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.4 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.3 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 0.3 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 0.3 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 0.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.3 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.9 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.8 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.6 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.2 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.1 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.4 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.1 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.0 | 0.2 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:2001188 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.0 | 0.2 | GO:2000506 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) positive regulation of eating behavior(GO:1904000) positive regulation of small intestine smooth muscle contraction(GO:1904349) negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 1.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 3.6 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.5 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.0 | 0.1 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.0 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 26.3 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0032202 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.0 | 0.5 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.5 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 1.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.2 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 1.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.0 | GO:0060305 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.0 | 0.2 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0043519 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 1.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 3.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.0 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.4 | 1.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.3 | 1.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 1.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.2 | 1.2 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.2 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0070401 | NADP+ binding(GO:0070401) lithocholic acid binding(GO:1902121) |
| 0.1 | 0.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.3 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.3 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.1 | 0.6 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.5 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.3 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 0.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.2 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 4.1 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 1.0 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 7.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 15.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 0.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.0 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.1 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |