Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
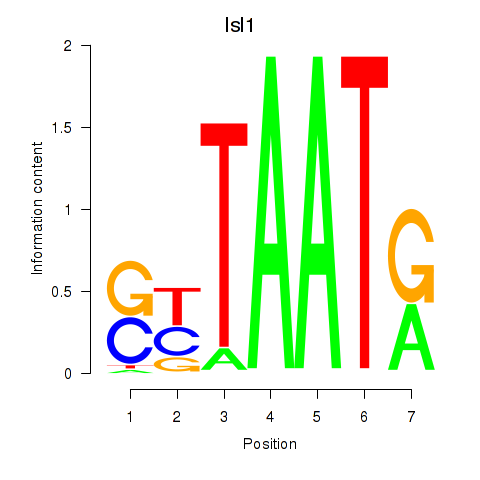
Results for Isl1
Z-value: 0.52
Transcription factors associated with Isl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl1
|
ENSMUSG00000042258.14 | ISL1 transcription factor, LIM/homeodomain |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl1 | mm39_v1_chr13_-_116446166_116446235 | -0.31 | 6.8e-02 | Click! |
Activity profile of Isl1 motif
Sorted Z-values of Isl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_140343652 | 3.90 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr1_+_67162176 | 2.33 |
ENSMUST00000027144.8
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr11_-_84058292 | 2.20 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr14_+_51333816 | 2.19 |
ENSMUST00000169895.3
|
Rnase4
|
ribonuclease, RNase A family 4 |
| chr19_+_4036562 | 1.89 |
ENSMUST00000236224.2
ENSMUST00000236510.2 ENSMUST00000237910.2 ENSMUST00000235612.2 ENSMUST00000054030.8 |
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr6_+_125298372 | 1.80 |
ENSMUST00000176442.8
ENSMUST00000177329.2 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr5_-_77262968 | 1.69 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr17_+_37253802 | 1.61 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr7_+_51537645 | 1.53 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr6_+_125298296 | 1.42 |
ENSMUST00000081440.14
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr5_+_90608751 | 1.41 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr3_-_107952146 | 1.26 |
ENSMUST00000178808.8
ENSMUST00000106670.2 ENSMUST00000029489.15 |
Gstm4
|
glutathione S-transferase, mu 4 |
| chr10_-_108846816 | 1.22 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr10_+_41395870 | 1.01 |
ENSMUST00000189300.2
|
Cd164
|
CD164 antigen |
| chr12_-_25147139 | 0.97 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr17_-_56424577 | 0.96 |
ENSMUST00000019808.12
|
Plin5
|
perilipin 5 |
| chr10_+_76411474 | 0.95 |
ENSMUST00000001183.8
|
Ftcd
|
formiminotransferase cyclodeaminase |
| chr1_+_133109059 | 0.93 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr3_-_131196213 | 0.89 |
ENSMUST00000197057.2
|
Sgms2
|
sphingomyelin synthase 2 |
| chr4_-_6275629 | 0.88 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr3_-_88460166 | 0.83 |
ENSMUST00000119002.2
ENSMUST00000029698.15 |
Lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr8_+_45960804 | 0.82 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr12_-_86931529 | 0.79 |
ENSMUST00000038422.8
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr19_+_8816663 | 0.75 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr4_-_141412910 | 0.72 |
ENSMUST00000105782.2
|
Rsc1a1
|
regulatory solute carrier protein, family 1, member 1 |
| chr5_+_90666791 | 0.65 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr18_+_77032080 | 0.63 |
ENSMUST00000026485.15
ENSMUST00000150990.9 ENSMUST00000148955.3 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr4_+_155045372 | 0.62 |
ENSMUST00000049621.7
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr17_-_35265514 | 0.62 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5 |
| chr10_-_107321938 | 0.61 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr5_-_104125192 | 0.60 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr8_-_62355690 | 0.60 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr8_+_93687561 | 0.59 |
ENSMUST00000072939.8
|
Slc6a2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| chr3_-_148696155 | 0.57 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr5_-_104125270 | 0.56 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr16_-_90866032 | 0.56 |
ENSMUST00000035689.8
ENSMUST00000114076.2 |
4932438H23Rik
|
RIKEN cDNA 4932438H23 gene |
| chr5_-_104125226 | 0.54 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr4_-_42168603 | 0.54 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr4_-_42665763 | 0.54 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr10_+_50770836 | 0.53 |
ENSMUST00000219436.2
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr3_-_75177378 | 0.52 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr17_+_46991972 | 0.51 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1 |
| chr6_+_125298168 | 0.50 |
ENSMUST00000176365.2
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr5_+_23992689 | 0.48 |
ENSMUST00000120869.6
ENSMUST00000030852.13 ENSMUST00000117783.8 ENSMUST00000115113.3 |
Rint1
|
RAD50 interactor 1 |
| chr17_-_35265702 | 0.47 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr14_-_36657517 | 0.47 |
ENSMUST00000183038.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr5_+_107655487 | 0.46 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr10_+_34359395 | 0.45 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr11_+_74540284 | 0.44 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr16_+_36514334 | 0.43 |
ENSMUST00000023617.13
ENSMUST00000089618.10 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr3_-_49711765 | 0.43 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr8_-_5155347 | 0.43 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr10_-_12490424 | 0.43 |
ENSMUST00000217994.2
|
Utrn
|
utrophin |
| chr1_-_162726053 | 0.42 |
ENSMUST00000143123.3
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr7_+_143792455 | 0.41 |
ENSMUST00000239495.2
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr6_+_113435716 | 0.41 |
ENSMUST00000203661.3
ENSMUST00000204774.3 ENSMUST00000053569.7 ENSMUST00000101065.8 |
Il17re
|
interleukin 17 receptor E |
| chr3_-_116388334 | 0.41 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
| chr9_+_21914083 | 0.40 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr17_-_24752683 | 0.40 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr2_+_67578556 | 0.40 |
ENSMUST00000180887.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr10_-_92917458 | 0.40 |
ENSMUST00000212902.2
ENSMUST00000168617.3 ENSMUST00000168110.8 ENSMUST00000020200.14 |
Cfap54
|
cilia and flagella associated protein 54 |
| chr4_+_42114817 | 0.40 |
ENSMUST00000098123.4
|
Gm13304
|
predicted gene 13304 |
| chr11_-_99412084 | 0.39 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr8_-_71964379 | 0.38 |
ENSMUST00000048452.6
|
Plvap
|
plasmalemma vesicle associated protein |
| chr3_-_49711706 | 0.38 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr8_-_34237752 | 0.38 |
ENSMUST00000179364.3
|
Smim18
|
small integral membrane protein 18 |
| chr4_+_122889407 | 0.38 |
ENSMUST00000144998.2
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr1_-_170695328 | 0.37 |
ENSMUST00000027974.7
|
Atf6
|
activating transcription factor 6 |
| chrX_-_103900834 | 0.37 |
ENSMUST00000033575.7
|
Magee2
|
MAGE family member E2 |
| chr10_+_34359513 | 0.36 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr2_+_153003212 | 0.36 |
ENSMUST00000089027.3
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr4_+_41903610 | 0.35 |
ENSMUST00000098128.4
|
Ccl21d
|
chemokine (C-C motif) ligand 21D |
| chr10_+_28544556 | 0.35 |
ENSMUST00000161345.2
|
Themis
|
thymocyte selection associated |
| chr5_-_87634665 | 0.34 |
ENSMUST00000201519.2
|
Gm43638
|
predicted gene 43638 |
| chr4_+_42255693 | 0.34 |
ENSMUST00000178864.3
|
Ccl21b
|
chemokine (C-C motif) ligand 21B (leucine) |
| chr15_-_101726600 | 0.33 |
ENSMUST00000023712.8
|
Krt2
|
keratin 2 |
| chr9_-_119038162 | 0.33 |
ENSMUST00000084797.6
|
Slc22a13
|
solute carrier family 22 (organic cation transporter), member 13 |
| chr10_+_102348076 | 0.33 |
ENSMUST00000219445.2
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chrX_+_71025128 | 0.33 |
ENSMUST00000114569.2
|
Fate1
|
fetal and adult testis expressed 1 |
| chr1_+_139349912 | 0.32 |
ENSMUST00000200243.5
ENSMUST00000039867.10 |
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr11_-_99412162 | 0.32 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr15_-_36496880 | 0.32 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr13_-_21726945 | 0.31 |
ENSMUST00000205976.3
ENSMUST00000175637.3 |
Olfr1366
|
olfactory receptor 1366 |
| chr14_+_67148619 | 0.31 |
ENSMUST00000089236.11
ENSMUST00000122431.3 |
Pnma2
|
paraneoplastic antigen MA2 |
| chr4_+_150321272 | 0.31 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr4_-_14621805 | 0.30 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr2_-_127498129 | 0.30 |
ENSMUST00000028853.7
|
Mal
|
myelin and lymphocyte protein, T cell differentiation protein |
| chr5_+_81169049 | 0.29 |
ENSMUST00000117253.8
ENSMUST00000120128.8 |
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr7_-_84059170 | 0.29 |
ENSMUST00000208995.2
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr4_-_14621669 | 0.29 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr11_-_121410152 | 0.29 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chrX_+_152020744 | 0.29 |
ENSMUST00000112574.9
|
Klf8
|
Kruppel-like factor 8 |
| chr3_-_123484499 | 0.29 |
ENSMUST00000154668.8
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr8_+_45960855 | 0.28 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_+_102927366 | 0.27 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr2_+_162785394 | 0.26 |
ENSMUST00000035751.12
ENSMUST00000156954.8 |
L3mbtl1
|
L3MBTL1 histone methyl-lysine binding protein |
| chr14_-_118289557 | 0.26 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr9_-_82856321 | 0.26 |
ENSMUST00000189985.2
|
Phip
|
pleckstrin homology domain interacting protein |
| chr5_-_108022900 | 0.26 |
ENSMUST00000138111.8
ENSMUST00000112642.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr16_-_94023976 | 0.25 |
ENSMUST00000227698.2
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase) |
| chr4_+_106954794 | 0.25 |
ENSMUST00000221740.2
|
Cdcp2
|
CUB domain containing protein 2 |
| chr18_-_35348049 | 0.25 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr16_+_33650002 | 0.24 |
ENSMUST00000115028.11
ENSMUST00000069345.6 |
Itgb5
|
integrin beta 5 |
| chrX_-_142716085 | 0.24 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr9_-_112016966 | 0.23 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr11_+_98277276 | 0.22 |
ENSMUST00000041301.8
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr10_+_28544356 | 0.21 |
ENSMUST00000060409.13
ENSMUST00000056097.11 ENSMUST00000105516.9 |
Themis
|
thymocyte selection associated |
| chr5_+_24679154 | 0.20 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr7_+_119289249 | 0.20 |
ENSMUST00000047045.10
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr5_+_121601212 | 0.19 |
ENSMUST00000094357.11
ENSMUST00000031405.12 |
Tmem116
|
transmembrane protein 116 |
| chr7_+_101879176 | 0.19 |
ENSMUST00000120119.9
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr15_+_102898966 | 0.18 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chr2_+_57887896 | 0.18 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr2_-_17465410 | 0.18 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr7_-_106354591 | 0.18 |
ENSMUST00000214306.2
ENSMUST00000216255.2 |
Olfr698
|
olfactory receptor 698 |
| chr2_+_85822163 | 0.18 |
ENSMUST00000050942.3
|
Olfr1031
|
olfactory receptor 1031 |
| chr4_+_52964547 | 0.18 |
ENSMUST00000215010.2
ENSMUST00000215127.2 |
Olfr270
|
olfactory receptor 270 |
| chr17_+_8384333 | 0.18 |
ENSMUST00000097419.10
ENSMUST00000024636.15 |
Cep43
|
centrosomal protein 43 |
| chr14_+_53081922 | 0.18 |
ENSMUST00000181360.3
ENSMUST00000183652.2 |
Trav12d-1
|
T cell receptor alpha variable 12D-1 |
| chr11_-_99313078 | 0.18 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr11_-_109188917 | 0.17 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr4_-_14621497 | 0.17 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr18_-_13074834 | 0.17 |
ENSMUST00000122175.8
ENSMUST00000142467.2 ENSMUST00000074352.11 |
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr6_-_144155197 | 0.16 |
ENSMUST00000038815.14
ENSMUST00000111749.8 ENSMUST00000170367.9 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr16_+_56298228 | 0.16 |
ENSMUST00000231832.2
ENSMUST00000096013.11 ENSMUST00000048471.15 ENSMUST00000231870.2 ENSMUST00000171000.3 ENSMUST00000231781.2 ENSMUST00000096012.11 |
Abi3bp
|
ABI family member 3 binding protein |
| chr8_-_41494890 | 0.16 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr7_+_114342929 | 0.15 |
ENSMUST00000161800.2
|
Insc
|
INSC spindle orientation adaptor protein |
| chr3_+_88461059 | 0.15 |
ENSMUST00000008748.8
|
Ubqln4
|
ubiquilin 4 |
| chr11_-_109188892 | 0.15 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr10_+_75983285 | 0.14 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr12_-_113945961 | 0.14 |
ENSMUST00000103466.2
|
Ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr4_-_126219434 | 0.14 |
ENSMUST00000131113.8
|
Tekt2
|
tektin 2 |
| chr8_+_70945806 | 0.13 |
ENSMUST00000008032.14
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr1_+_87731360 | 0.13 |
ENSMUST00000177757.2
ENSMUST00000077772.12 |
Sag
|
S-antigen, retina and pineal gland (arrestin) |
| chr3_-_7678785 | 0.13 |
ENSMUST00000194279.6
|
Il7
|
interleukin 7 |
| chr15_-_34356567 | 0.12 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr17_-_71153283 | 0.12 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr2_-_89671899 | 0.12 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr8_+_94993453 | 0.11 |
ENSMUST00000212167.2
|
Nup93
|
nucleoporin 93 |
| chr11_+_117157024 | 0.11 |
ENSMUST00000019038.15
|
Septin9
|
septin 9 |
| chr14_-_121976273 | 0.11 |
ENSMUST00000212376.3
ENSMUST00000239077.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr9_-_112046411 | 0.11 |
ENSMUST00000161412.8
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr10_-_102864904 | 0.11 |
ENSMUST00000167156.9
|
Alx1
|
ALX homeobox 1 |
| chr6_-_144155167 | 0.11 |
ENSMUST00000077160.12
|
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr14_+_53167246 | 0.11 |
ENSMUST00000177703.3
|
Trav12d-3
|
T cell receptor alpha variable 12D-3 |
| chrX_-_142716200 | 0.10 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr2_-_86882616 | 0.10 |
ENSMUST00000213781.2
ENSMUST00000217650.2 |
Olfr1106
|
olfactory receptor 1106 |
| chr8_+_11847411 | 0.10 |
ENSMUST00000239427.2
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr3_-_144511566 | 0.10 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chrX_-_151820545 | 0.10 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr17_-_35454729 | 0.10 |
ENSMUST00000048994.7
|
Nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor like 1 |
| chr12_-_73093953 | 0.10 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr1_-_58009263 | 0.09 |
ENSMUST00000114410.10
|
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr10_-_130050272 | 0.09 |
ENSMUST00000213568.2
|
Olfr827
|
olfactory receptor 827 |
| chrX_+_7446721 | 0.09 |
ENSMUST00000115738.8
|
Foxp3
|
forkhead box P3 |
| chr11_-_6180127 | 0.09 |
ENSMUST00000004505.3
|
Npc1l1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr11_+_46641330 | 0.09 |
ENSMUST00000109223.8
|
Havcr1
|
hepatitis A virus cellular receptor 1 |
| chr5_+_129661233 | 0.08 |
ENSMUST00000031390.10
|
Mmp17
|
matrix metallopeptidase 17 |
| chr7_-_102241093 | 0.08 |
ENSMUST00000213540.3
|
Olfr551
|
olfactory receptor 551 |
| chr1_+_81055201 | 0.08 |
ENSMUST00000123285.2
|
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr4_+_57821050 | 0.08 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr2_+_111491764 | 0.08 |
ENSMUST00000213511.2
ENSMUST00000207228.3 ENSMUST00000208983.2 |
Olfr1299
Gm44840
|
olfactory receptor 1299 predicted gene 44840 |
| chr18_-_15851103 | 0.07 |
ENSMUST00000053017.13
|
Chst9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr13_-_24175141 | 0.07 |
ENSMUST00000021770.8
|
Scgn
|
secretagogin, EF-hand calcium binding protein |
| chr10_+_18345706 | 0.07 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr10_-_102864921 | 0.07 |
ENSMUST00000217946.2
ENSMUST00000219194.2 |
Alx1
|
ALX homeobox 1 |
| chr14_+_54457978 | 0.07 |
ENSMUST00000103740.2
ENSMUST00000198398.5 |
Trac
|
T cell receptor alpha constant |
| chr6_-_54949587 | 0.07 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chrX_+_113384008 | 0.07 |
ENSMUST00000113371.8
ENSMUST00000040504.12 |
Klhl4
|
kelch-like 4 |
| chr3_-_37180093 | 0.07 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr10_-_70491764 | 0.07 |
ENSMUST00000162144.2
ENSMUST00000162793.8 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr14_-_50425655 | 0.07 |
ENSMUST00000205837.3
|
Olfr730
|
olfactory receptor 730 |
| chr14_+_53775648 | 0.06 |
ENSMUST00000200115.2
ENSMUST00000103650.3 |
Trav12-1
|
T cell receptor alpha variable 12-1 |
| chr6_+_54794433 | 0.06 |
ENSMUST00000127331.2
|
Znrf2
|
zinc and ring finger 2 |
| chr19_+_12839106 | 0.05 |
ENSMUST00000059675.4
|
Olfr1444
|
olfactory receptor 1444 |
| chr3_-_7678796 | 0.05 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr2_-_120946877 | 0.05 |
ENSMUST00000110675.3
|
Tgm7
|
transglutaminase 7 |
| chr9_+_94551929 | 0.05 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr9_+_37278647 | 0.05 |
ENSMUST00000051839.9
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chrX_+_141882590 | 0.04 |
ENSMUST00000165829.3
|
Rtl9
|
retrotransposon Gag like 9 |
| chr16_-_58583539 | 0.04 |
ENSMUST00000214139.2
|
Olfr172
|
olfactory receptor 172 |
| chr9_+_3403592 | 0.04 |
ENSMUST00000027027.7
|
Cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr14_-_96756503 | 0.04 |
ENSMUST00000022666.9
|
Klhl1
|
kelch-like 1 |
| chr16_-_96971905 | 0.04 |
ENSMUST00000056102.9
|
Dscam
|
DS cell adhesion molecule |
| chr14_+_47535717 | 0.04 |
ENSMUST00000166743.9
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr9_-_44679136 | 0.04 |
ENSMUST00000034607.10
|
Arcn1
|
archain 1 |
| chr14_-_78866714 | 0.04 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr4_+_43406435 | 0.04 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr6_+_90105406 | 0.04 |
ENSMUST00000072859.6
|
Vmn1r51
|
vomeronasal 1 receptor 51 |
| chr5_+_149452144 | 0.04 |
ENSMUST00000110502.7
|
Wdr95
|
WD40 repeat domain 95 |
| chr6_-_129428869 | 0.04 |
ENSMUST00000203162.3
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr11_-_99884818 | 0.04 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr4_+_41966058 | 0.03 |
ENSMUST00000108026.3
|
Fam205a4
|
family with sequence similarity 205, member A4 |
| chr19_-_11816583 | 0.03 |
ENSMUST00000214887.2
|
Olfr1417
|
olfactory receptor 1417 |
| chr5_+_20112704 | 0.03 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_+_73931679 | 0.03 |
ENSMUST00000098006.9
ENSMUST00000084474.6 |
Frmd3
|
FERM domain containing 3 |
| chr6_+_30568366 | 0.03 |
ENSMUST00000049251.6
|
Cpa4
|
carboxypeptidase A4 |
| chr17_+_71511642 | 0.03 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr2_+_71042285 | 0.03 |
ENSMUST00000112144.9
ENSMUST00000100028.10 ENSMUST00000112136.2 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr1_-_79417732 | 0.02 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.3 | 2.3 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.3 | 1.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.6 | GO:2000978 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.2 | 1.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.9 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.2 | 1.3 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.2 | 0.9 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.2 | 0.5 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.2 | 4.3 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 0.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 3.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.3 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.1 | 0.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.7 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.8 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.4 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0002660 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.6 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.6 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 1.1 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 1.7 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 0.5 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 3.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.8 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 1.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 2.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 5.9 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.4 | 1.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 3.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 1.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 0.6 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 0.6 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 0.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.4 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 3.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 1.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.0 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.3 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 2.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.7 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 1.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |