Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
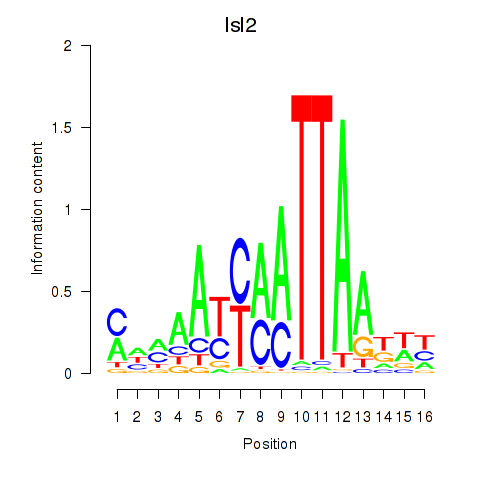
Results for Isl2
Z-value: 0.46
Transcription factors associated with Isl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl2
|
ENSMUSG00000032318.13 | insulin related protein 2 (islet 2) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl2 | mm39_v1_chr9_+_55448432_55448493 | -0.38 | 2.2e-02 | Click! |
Activity profile of Isl2 motif
Sorted Z-values of Isl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_114860375 | 3.49 |
ENSMUST00000147605.8
ENSMUST00000134593.2 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr6_-_41012435 | 3.32 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr17_-_28779678 | 3.14 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr8_+_114860342 | 2.95 |
ENSMUST00000109109.8
|
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr8_+_114860297 | 2.69 |
ENSMUST00000073521.12
ENSMUST00000066514.13 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr19_-_39637489 | 2.48 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr19_-_8109346 | 2.03 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr19_+_12673147 | 2.00 |
ENSMUST00000025598.10
ENSMUST00000138545.8 ENSMUST00000154822.2 |
Keg1
|
kidney expressed gene 1 |
| chr3_-_67422821 | 1.98 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr1_+_88139678 | 1.93 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr1_-_139786421 | 1.56 |
ENSMUST00000194186.6
ENSMUST00000094489.5 ENSMUST00000239380.2 |
Cfhr2
|
complement factor H-related 2 |
| chr17_-_56312555 | 1.46 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr6_-_136899167 | 1.32 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr19_-_39801188 | 1.27 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr1_-_121260298 | 1.17 |
ENSMUST00000071064.13
|
Insig2
|
insulin induced gene 2 |
| chr19_-_39875192 | 1.13 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr9_+_118931532 | 1.13 |
ENSMUST00000165231.8
ENSMUST00000140326.8 |
Dlec1
|
deleted in lung and esophageal cancer 1 |
| chr19_+_37425180 | 1.12 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr1_+_87983099 | 1.11 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr1_+_87998487 | 1.09 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr10_+_23727325 | 1.08 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr7_-_100306160 | 1.05 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr4_+_43493344 | 0.89 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr17_+_85335775 | 0.87 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr1_-_121260274 | 0.83 |
ENSMUST00000161068.2
|
Insig2
|
insulin induced gene 2 |
| chr5_+_87148697 | 0.81 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr1_+_87983189 | 0.80 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr1_+_180878797 | 0.76 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chrM_+_10167 | 0.71 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_+_89366425 | 0.71 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr3_-_75177378 | 0.71 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr5_+_90708962 | 0.70 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chrX_+_100427331 | 0.68 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr13_-_12476313 | 0.67 |
ENSMUST00000143693.8
ENSMUST00000144283.2 ENSMUST00000099820.10 ENSMUST00000135166.8 |
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr9_-_15212849 | 0.65 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr9_+_119170486 | 0.65 |
ENSMUST00000175743.8
ENSMUST00000176397.8 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr7_+_86895996 | 0.63 |
ENSMUST00000068829.13
|
Nox4
|
NADPH oxidase 4 |
| chr7_-_12829100 | 0.61 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr4_-_119217079 | 0.61 |
ENSMUST00000143494.3
ENSMUST00000154606.9 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr11_+_109434519 | 0.60 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr11_+_5470652 | 0.58 |
ENSMUST00000063084.16
|
Xbp1
|
X-box binding protein 1 |
| chr9_+_108167628 | 0.57 |
ENSMUST00000035227.8
|
Nicn1
|
nicolin 1 |
| chrM_+_9459 | 0.56 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr6_-_83098255 | 0.56 |
ENSMUST00000205023.2
ENSMUST00000146328.4 ENSMUST00000151393.7 ENSMUST00000032111.11 ENSMUST00000113936.10 |
Wbp1
|
WW domain binding protein 1 |
| chr9_+_107765320 | 0.55 |
ENSMUST00000191906.6
ENSMUST00000035202.4 |
Mon1a
|
MON1 homolog A, secretory traffciking associated |
| chr11_-_101169753 | 0.52 |
ENSMUST00000168089.2
ENSMUST00000017332.4 |
Coa3
|
cytochrome C oxidase assembly factor 3 |
| chr6_-_115569504 | 0.47 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr6_+_124281607 | 0.46 |
ENSMUST00000032234.5
ENSMUST00000112541.8 |
Cd163
|
CD163 antigen |
| chrM_+_3906 | 0.40 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr8_+_127898139 | 0.39 |
ENSMUST00000238870.2
|
Pard3
|
par-3 family cell polarity regulator |
| chr17_+_79919267 | 0.38 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chrM_+_14138 | 0.38 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr4_+_22357543 | 0.38 |
ENSMUST00000039234.10
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr12_-_84664001 | 0.37 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chrM_+_9870 | 0.37 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr6_+_78347636 | 0.37 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr5_-_3697806 | 0.35 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr11_+_94218810 | 0.35 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr11_-_49004584 | 0.35 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr3_-_96104886 | 0.34 |
ENSMUST00000016087.4
|
Bola1
|
bolA-like 1 (E. coli) |
| chr11_+_94219046 | 0.34 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr19_+_45064539 | 0.33 |
ENSMUST00000026234.5
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr15_+_100251030 | 0.33 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr2_+_69727563 | 0.33 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr5_-_118382926 | 0.33 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr2_+_68966125 | 0.32 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chr12_+_10440755 | 0.32 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr9_-_51240201 | 0.32 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr9_+_107440484 | 0.32 |
ENSMUST00000193418.2
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr1_-_57008986 | 0.32 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr13_-_43634695 | 0.31 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr11_+_69729340 | 0.31 |
ENSMUST00000133967.8
ENSMUST00000094065.5 |
Tmem256
|
transmembrane protein 256 |
| chr15_+_99192968 | 0.31 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr6_+_40619913 | 0.30 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr9_+_21634779 | 0.30 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr3_-_49711706 | 0.30 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr16_-_48592319 | 0.30 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chrX_-_103024847 | 0.30 |
ENSMUST00000121153.8
ENSMUST00000070705.6 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr13_+_74554509 | 0.29 |
ENSMUST00000222435.2
|
Ftl1-ps1
|
ferritin light polypeptide 1, pseudogene 1 |
| chr4_+_105014536 | 0.29 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr19_-_7943365 | 0.28 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr11_+_58062467 | 0.28 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr9_+_81745723 | 0.28 |
ENSMUST00000057067.10
ENSMUST00000189832.7 ENSMUST00000189391.2 |
Mei4
|
meiotic double-stranded break formation protein 4 |
| chr6_+_134617903 | 0.27 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr3_-_49711765 | 0.27 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr15_-_3333003 | 0.27 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr6_+_78382131 | 0.26 |
ENSMUST00000023906.4
|
Reg2
|
regenerating islet-derived 2 |
| chr6_-_108162513 | 0.25 |
ENSMUST00000167338.8
ENSMUST00000172188.2 ENSMUST00000032191.16 |
Sumf1
|
sulfatase modifying factor 1 |
| chr9_-_15212745 | 0.24 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr17_-_40630096 | 0.24 |
ENSMUST00000026498.5
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr14_-_68771138 | 0.24 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr8_+_72973560 | 0.24 |
ENSMUST00000003123.10
|
Fam32a
|
family with sequence similarity 32, member A |
| chr16_-_48592372 | 0.23 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr8_+_121842902 | 0.23 |
ENSMUST00000054691.8
|
Foxc2
|
forkhead box C2 |
| chr2_+_124978518 | 0.23 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr16_+_11224481 | 0.23 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr15_+_79113341 | 0.22 |
ENSMUST00000163571.8
|
Pick1
|
protein interacting with C kinase 1 |
| chr7_+_18853778 | 0.21 |
ENSMUST00000053109.5
|
Fbxo46
|
F-box protein 46 |
| chr3_-_94949854 | 0.21 |
ENSMUST00000117355.2
ENSMUST00000071664.12 ENSMUST00000107237.8 |
Psmd4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| chr1_+_155911136 | 0.21 |
ENSMUST00000111757.10
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr19_+_43741550 | 0.21 |
ENSMUST00000153295.2
|
Cutc
|
cutC copper transporter |
| chr11_+_97732108 | 0.21 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr4_-_14621669 | 0.21 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr5_+_88073483 | 0.20 |
ENSMUST00000113271.3
|
Csn3
|
casein kappa |
| chr12_+_86993805 | 0.20 |
ENSMUST00000189246.7
|
Cipc
|
CLOCK interacting protein, circadian |
| chr2_+_69727599 | 0.19 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_+_127696548 | 0.19 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr8_-_55177352 | 0.19 |
ENSMUST00000129132.3
ENSMUST00000150488.8 ENSMUST00000127511.9 |
Wdr17
|
WD repeat domain 17 |
| chr4_-_14621805 | 0.19 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr17_+_26934617 | 0.19 |
ENSMUST00000062519.14
ENSMUST00000144221.2 ENSMUST00000142539.8 ENSMUST00000151681.2 |
Crebrf
|
CREB3 regulatory factor |
| chr4_-_94817025 | 0.19 |
ENSMUST00000030309.6
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr13_+_67981349 | 0.19 |
ENSMUST00000222626.2
ENSMUST00000060609.8 |
Gm10037
|
predicted gene 10037 |
| chr14_+_70077841 | 0.19 |
ENSMUST00000022678.5
|
Pebp4
|
phosphatidylethanolamine binding protein 4 |
| chr6_+_125026865 | 0.18 |
ENSMUST00000112413.8
|
Acrbp
|
proacrosin binding protein |
| chr17_+_79223946 | 0.17 |
ENSMUST00000063817.7
|
Cebpzos
|
CCAAT/enhancer binding protein (C/EBP), zeta, opposite strand |
| chr2_+_36575800 | 0.17 |
ENSMUST00000213258.2
|
Olfr346
|
olfactory receptor 346 |
| chr6_+_65019793 | 0.17 |
ENSMUST00000204696.3
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr19_-_44541270 | 0.17 |
ENSMUST00000166808.2
|
Gm20538
|
predicted gene 20538 |
| chr9_-_102231884 | 0.17 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chr9_+_100479732 | 0.17 |
ENSMUST00000124487.8
|
Stag1
|
stromal antigen 1 |
| chr1_-_160405464 | 0.17 |
ENSMUST00000238289.2
|
Gpr52
|
G protein-coupled receptor 52 |
| chr2_+_87854404 | 0.17 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr14_-_54520382 | 0.16 |
ENSMUST00000059996.7
|
Olfr49
|
olfactory receptor 49 |
| chr18_-_73887528 | 0.16 |
ENSMUST00000041138.3
|
Elac1
|
elaC ribonuclease Z 1 |
| chr5_+_107655487 | 0.16 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr2_+_124978612 | 0.16 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr4_-_86587728 | 0.16 |
ENSMUST00000149700.8
|
Plin2
|
perilipin 2 |
| chr14_+_122272069 | 0.16 |
ENSMUST00000045976.7
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chr16_-_17379103 | 0.16 |
ENSMUST00000232637.2
ENSMUST00000100123.10 ENSMUST00000023442.13 |
Lrrc74b
|
leucine rich repeat containing 74B |
| chr3_+_41517790 | 0.16 |
ENSMUST00000163764.8
|
Jade1
|
jade family PHD finger 1 |
| chr14_+_53574579 | 0.16 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chrM_-_14061 | 0.15 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr1_+_88234454 | 0.15 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chrX_-_111315519 | 0.15 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr5_+_14564932 | 0.15 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr12_+_116239006 | 0.15 |
ENSMUST00000090195.5
|
Gm11027
|
predicted gene 11027 |
| chr9_-_123461593 | 0.15 |
ENSMUST00000026273.11
|
Slc6a20b
|
solute carrier family 6 (neurotransmitter transporter), member 20B |
| chr5_+_87814058 | 0.15 |
ENSMUST00000199506.5
ENSMUST00000197631.5 ENSMUST00000094641.9 |
Csn1s1
|
casein alpha s1 |
| chr14_-_63221950 | 0.15 |
ENSMUST00000100493.3
|
Defb48
|
defensin beta 48 |
| chr4_+_102843540 | 0.14 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr17_+_14087827 | 0.14 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr13_-_67599702 | 0.14 |
ENSMUST00000225479.2
ENSMUST00000057241.15 ENSMUST00000075255.6 |
Zfp874a
|
zinc finger protein 874a |
| chr9_+_18320390 | 0.14 |
ENSMUST00000098973.3
|
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr13_+_23343482 | 0.14 |
ENSMUST00000226845.2
ENSMUST00000228666.2 ENSMUST00000227388.2 |
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chrX_+_72719098 | 0.14 |
ENSMUST00000171398.2
|
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr13_+_22563988 | 0.13 |
ENSMUST00000227685.2
ENSMUST00000227689.2 ENSMUST00000227846.2 |
Vmn1r199
|
vomeronasal 1 receptor 199 |
| chr5_+_88073438 | 0.13 |
ENSMUST00000001667.13
ENSMUST00000113267.8 |
Csn3
|
casein kappa |
| chr9_+_121606750 | 0.13 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40 |
| chr10_+_61007733 | 0.13 |
ENSMUST00000122261.8
ENSMUST00000121297.8 ENSMUST00000035894.12 |
Tbata
|
thymus, brain and testes associated |
| chr8_-_62355690 | 0.13 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr16_-_44153288 | 0.13 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr1_+_44187007 | 0.13 |
ENSMUST00000027214.4
|
Ercc5
|
excision repair cross-complementing rodent repair deficiency, complementation group 5 |
| chr7_-_19621833 | 0.13 |
ENSMUST00000052605.8
|
Ceacam19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr9_-_107749594 | 0.13 |
ENSMUST00000183035.2
|
Rbm6
|
RNA binding motif protein 6 |
| chr14_-_51433380 | 0.12 |
ENSMUST00000051274.2
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr1_-_169938298 | 0.12 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr13_+_19394484 | 0.12 |
ENSMUST00000200495.2
|
Trgj1
|
T cell receptor gamma joining 1 |
| chr15_-_35938155 | 0.12 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr7_-_106392405 | 0.12 |
ENSMUST00000215952.2
ENSMUST00000216307.2 |
Olfr699
|
olfactory receptor 699 |
| chrM_+_8603 | 0.12 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr5_-_25107313 | 0.12 |
ENSMUST00000131486.2
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr11_-_97909134 | 0.11 |
ENSMUST00000107561.9
|
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr14_+_65504067 | 0.11 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr8_+_22682816 | 0.11 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr2_-_17465410 | 0.11 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr1_-_139304832 | 0.11 |
ENSMUST00000198445.5
|
Crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr3_+_87343071 | 0.11 |
ENSMUST00000194102.6
|
Fcrl5
|
Fc receptor-like 5 |
| chr4_+_147592057 | 0.11 |
ENSMUST00000063704.8
|
Zfp982
|
zinc finger protein 982 |
| chr4_-_101838246 | 0.10 |
ENSMUST00000106916.2
|
Gm12790
|
predicted gene 12790 |
| chr3_+_87343127 | 0.10 |
ENSMUST00000166297.7
ENSMUST00000049926.15 ENSMUST00000178261.3 |
Fcrl5
|
Fc receptor-like 5 |
| chr3_+_87343105 | 0.10 |
ENSMUST00000193229.6
|
Fcrl5
|
Fc receptor-like 5 |
| chr1_+_173876771 | 0.10 |
ENSMUST00000213381.2
ENSMUST00000213211.2 |
Olfr432
|
olfactory receptor 432 |
| chr4_-_14621497 | 0.10 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chrY_-_1245685 | 0.10 |
ENSMUST00000143286.8
ENSMUST00000137048.8 ENSMUST00000069309.14 ENSMUST00000139365.8 ENSMUST00000154004.8 |
Uty
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr5_+_73164226 | 0.10 |
ENSMUST00000031127.11
ENSMUST00000201304.2 |
Slc10a4
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 4 |
| chr1_+_46105898 | 0.10 |
ENSMUST00000069293.10
|
Dnah7b
|
dynein, axonemal, heavy chain 7B |
| chr7_-_106354591 | 0.10 |
ENSMUST00000214306.2
ENSMUST00000216255.2 |
Olfr698
|
olfactory receptor 698 |
| chr19_+_4015900 | 0.09 |
ENSMUST00000025794.7
|
Aldh3b3
|
aldehyde dehydrogenase 3 family, member B3 |
| chr10_-_128885867 | 0.09 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr10_+_29074950 | 0.09 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr8_-_65582206 | 0.09 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr2_+_85876205 | 0.09 |
ENSMUST00000213496.2
|
Olfr1034
|
olfactory receptor 1034 |
| chr11_+_58485940 | 0.09 |
ENSMUST00000214990.2
ENSMUST00000216965.2 |
Olfr324
|
olfactory receptor 324 |
| chr7_-_106531426 | 0.09 |
ENSMUST00000215468.2
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr2_+_57887896 | 0.09 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr7_+_126575781 | 0.09 |
ENSMUST00000206450.2
ENSMUST00000205830.2 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr6_+_67873135 | 0.09 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chrX_+_163221035 | 0.09 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr14_+_53562089 | 0.09 |
ENSMUST00000178100.3
|
Trav7n-6
|
T cell receptor alpha variable 7N-6 |
| chr4_-_94817056 | 0.09 |
ENSMUST00000107097.9
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr13_+_22508759 | 0.09 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr4_+_139380849 | 0.09 |
ENSMUST00000030510.14
ENSMUST00000166773.2 |
Tas1r2
|
taste receptor, type 1, member 2 |
| chr7_-_103191924 | 0.09 |
ENSMUST00000214269.3
|
Olfr612
|
olfactory receptor 612 |
| chr2_+_36263531 | 0.08 |
ENSMUST00000072114.4
ENSMUST00000217511.2 |
Olfr338
|
olfactory receptor 338 |
| chr8_+_96429665 | 0.08 |
ENSMUST00000073139.14
ENSMUST00000080666.8 ENSMUST00000212160.2 |
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr4_+_146033882 | 0.08 |
ENSMUST00000105730.2
ENSMUST00000091878.6 |
Zfp987
|
zinc finger protein 987 |
| chr6_-_70313491 | 0.08 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr7_-_139982831 | 0.08 |
ENSMUST00000080153.4
ENSMUST00000216053.2 ENSMUST00000217167.2 |
Olfr531
|
olfactory receptor 531 |
| chr3_+_84081411 | 0.08 |
ENSMUST00000193882.2
|
Gm6525
|
predicted pseudogene 6525 |
| chr18_-_36899245 | 0.08 |
ENSMUST00000061522.8
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.1 | GO:0033869 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.6 | 1.9 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.3 | 3.0 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.6 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.2 | 1.1 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.2 | 0.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 4.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 2.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 2.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.3 | GO:0052572 | interleukin-15 production(GO:0032618) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.1 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.3 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 3.1 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.7 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.3 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.3 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0015881 | creatine transport(GO:0015881) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 1.1 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.3 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.0 | 0.2 | GO:2001015 | optic nerve morphogenesis(GO:0021631) negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.6 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.0 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 10.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.4 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.0 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.5 | 2.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 0.6 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.2 | 0.8 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 5.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 4.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 2.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.3 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 3.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.7 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 1.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 3.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |