Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
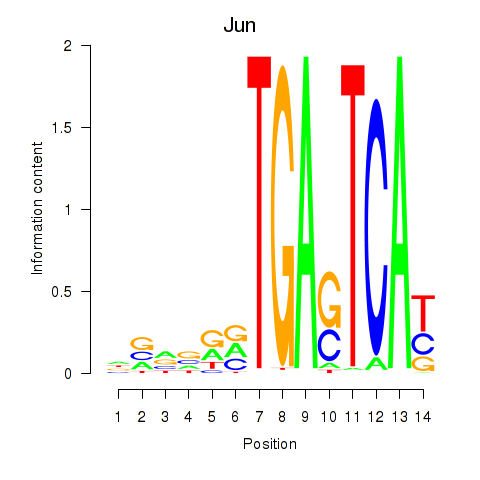
Results for Jun
Z-value: 0.86
Transcription factors associated with Jun
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Jun
|
ENSMUSG00000052684.5 | jun proto-oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Jun | mm39_v1_chr4_-_94940425_94940459 | -0.04 | 8.4e-01 | Click! |
Activity profile of Jun motif
Sorted Z-values of Jun motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_140856642 | 6.78 |
ENSMUST00000080654.7
ENSMUST00000167263.9 |
Cdhr5
|
cadherin-related family member 5 |
| chr9_-_86577940 | 6.69 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr12_-_103739847 | 4.08 |
ENSMUST00000078869.6
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr14_+_66205932 | 4.04 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr1_-_180021039 | 3.95 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr12_-_103923145 | 3.84 |
ENSMUST00000085054.5
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr12_-_103871146 | 3.79 |
ENSMUST00000074051.6
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr19_-_4087907 | 3.73 |
ENSMUST00000237982.2
|
Gstp1
|
glutathione S-transferase, pi 1 |
| chr19_-_4087940 | 3.67 |
ENSMUST00000237893.2
ENSMUST00000169613.4 |
Gstp1
|
glutathione S-transferase, pi 1 |
| chr1_+_133291302 | 3.50 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr19_-_4092218 | 3.48 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr7_-_28947882 | 3.47 |
ENSMUST00000032808.6
|
2200002D01Rik
|
RIKEN cDNA 2200002D01 gene |
| chr2_+_162829250 | 3.16 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr2_+_162829422 | 3.12 |
ENSMUST00000117123.2
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr18_+_12637217 | 2.97 |
ENSMUST00000188815.2
|
Lama3
|
laminin, alpha 3 |
| chr11_+_100973391 | 2.89 |
ENSMUST00000001806.10
ENSMUST00000107308.4 |
Coasy
|
Coenzyme A synthase |
| chr6_+_129510331 | 2.81 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr5_-_87402659 | 2.65 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr7_+_30252687 | 2.57 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr1_-_180021218 | 2.56 |
ENSMUST00000159914.8
|
Coq8a
|
coenzyme Q8A |
| chr7_+_140918793 | 2.56 |
ENSMUST00000026577.13
|
Eps8l2
|
EPS8-like 2 |
| chr7_+_127400016 | 2.49 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr12_-_103704417 | 2.48 |
ENSMUST00000095450.11
ENSMUST00000187220.2 |
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr12_-_103829810 | 2.48 |
ENSMUST00000085056.8
ENSMUST00000072876.12 ENSMUST00000124717.2 |
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
| chr2_-_25351106 | 2.46 |
ENSMUST00000114261.9
|
Paxx
|
non-homologous end joining factor |
| chr4_+_63274388 | 2.40 |
ENSMUST00000006687.5
|
Orm3
|
orosomucoid 3 |
| chr1_+_165596961 | 2.35 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr6_+_129510117 | 2.28 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr17_+_85335775 | 2.27 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr10_+_128769642 | 2.26 |
ENSMUST00000099112.4
ENSMUST00000218290.2 |
Itga7
|
integrin alpha 7 |
| chr16_+_90017634 | 2.23 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr2_-_25351024 | 2.19 |
ENSMUST00000151239.2
|
Paxx
|
non-homologous end joining factor |
| chr7_-_80051455 | 2.16 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr5_-_66238313 | 2.11 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr6_-_3968365 | 2.06 |
ENSMUST00000031674.11
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr2_+_143757193 | 2.02 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr18_+_21077627 | 2.01 |
ENSMUST00000050004.3
|
Rnf125
|
ring finger protein 125 |
| chr9_-_106353792 | 1.98 |
ENSMUST00000214682.2
ENSMUST00000112479.9 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr11_-_21521934 | 1.92 |
ENSMUST00000239073.2
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chrX_+_7588505 | 1.92 |
ENSMUST00000207675.2
ENSMUST00000116633.9 ENSMUST00000208996.2 ENSMUST00000144148.4 ENSMUST00000125991.9 ENSMUST00000148624.8 |
Wdr45
|
WD repeat domain 45 |
| chr17_-_24217068 | 1.91 |
ENSMUST00000041649.8
|
Prss22
|
protease, serine 22 |
| chr2_+_69210775 | 1.87 |
ENSMUST00000063690.4
|
Dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_+_182392559 | 1.82 |
ENSMUST00000168514.7
|
Capn8
|
calpain 8 |
| chr3_+_137923521 | 1.80 |
ENSMUST00000090171.7
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_-_103423472 | 1.80 |
ENSMUST00000044687.7
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr18_-_35760260 | 1.78 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chrX_+_7588453 | 1.77 |
ENSMUST00000043045.10
ENSMUST00000207386.2 ENSMUST00000116634.9 ENSMUST00000208072.2 ENSMUST00000207589.2 ENSMUST00000208618.2 ENSMUST00000208443.2 ENSMUST00000207541.2 ENSMUST00000208528.2 ENSMUST00000115689.10 ENSMUST00000131077.9 ENSMUST00000115688.8 ENSMUST00000208156.2 |
Wdr45
Gm45208
|
WD repeat domain 45 predicted gene 45208 |
| chr9_-_106353571 | 1.77 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr7_-_4973960 | 1.74 |
ENSMUST00000144863.8
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr14_+_66872699 | 1.72 |
ENSMUST00000159365.8
ENSMUST00000054661.8 ENSMUST00000225182.2 ENSMUST00000159068.2 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr3_-_131196213 | 1.71 |
ENSMUST00000197057.2
|
Sgms2
|
sphingomyelin synthase 2 |
| chr7_+_140918876 | 1.71 |
ENSMUST00000143633.4
|
Eps8l2
|
EPS8-like 2 |
| chr10_+_18720760 | 1.70 |
ENSMUST00000019998.9
|
Perp
|
PERP, TP53 apoptosis effector |
| chr11_-_50101592 | 1.64 |
ENSMUST00000143379.2
ENSMUST00000015981.12 ENSMUST00000102774.11 |
Sqstm1
|
sequestosome 1 |
| chr15_-_11399666 | 1.64 |
ENSMUST00000022849.7
|
Tars
|
threonyl-tRNA synthetase |
| chr11_+_87482971 | 1.62 |
ENSMUST00000103179.10
ENSMUST00000092802.12 ENSMUST00000146871.8 |
Mtmr4
|
myotubularin related protein 4 |
| chr2_+_151947444 | 1.62 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr13_+_30843937 | 1.60 |
ENSMUST00000091672.13
|
Dusp22
|
dual specificity phosphatase 22 |
| chr8_-_110766009 | 1.58 |
ENSMUST00000212934.2
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr8_-_5155347 | 1.56 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr13_+_30844025 | 1.55 |
ENSMUST00000110310.9
ENSMUST00000095914.7 |
Dusp22
|
dual specificity phosphatase 22 |
| chr7_-_133384449 | 1.52 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr7_-_127545896 | 1.48 |
ENSMUST00000118755.8
ENSMUST00000094026.10 |
Prss36
|
protease, serine 36 |
| chr7_-_44320244 | 1.47 |
ENSMUST00000048102.15
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr5_-_148489457 | 1.46 |
ENSMUST00000079324.14
|
Ubl3
|
ubiquitin-like 3 |
| chr7_+_24310171 | 1.43 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr15_-_75886166 | 1.43 |
ENSMUST00000060807.12
|
Fam83h
|
family with sequence similarity 83, member H |
| chr7_-_4974167 | 1.43 |
ENSMUST00000133272.2
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr14_-_34032311 | 1.40 |
ENSMUST00000111917.3
ENSMUST00000228704.2 |
Shld2
|
shieldin complex subunit 2 |
| chr3_+_121517158 | 1.37 |
ENSMUST00000029771.13
|
F3
|
coagulation factor III |
| chr1_+_182392577 | 1.36 |
ENSMUST00000048941.14
|
Capn8
|
calpain 8 |
| chr5_-_148489593 | 1.35 |
ENSMUST00000201595.4
ENSMUST00000164904.2 |
Ubl3
|
ubiquitin-like 3 |
| chr7_+_139414057 | 1.34 |
ENSMUST00000026548.14
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr12_-_85335193 | 1.33 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr6_+_17463748 | 1.31 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr15_-_4008913 | 1.29 |
ENSMUST00000022791.9
|
Fbxo4
|
F-box protein 4 |
| chr14_-_34032450 | 1.25 |
ENSMUST00000227375.2
|
Shld2
|
shieldin complex subunit 2 |
| chr8_-_110765983 | 1.23 |
ENSMUST00000109222.4
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr11_-_99213769 | 1.23 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr4_-_140393185 | 1.21 |
ENSMUST00000069623.12
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr19_+_8966641 | 1.20 |
ENSMUST00000092956.4
ENSMUST00000092955.11 |
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr16_-_56537808 | 1.19 |
ENSMUST00000065515.14
|
Tfg
|
Trk-fused gene |
| chr11_-_100139728 | 1.17 |
ENSMUST00000007280.9
|
Krt16
|
keratin 16 |
| chr7_-_126183392 | 1.17 |
ENSMUST00000128970.8
ENSMUST00000116269.9 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr1_-_175319842 | 1.16 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr17_+_53786240 | 1.16 |
ENSMUST00000017975.7
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr14_+_120715855 | 1.13 |
ENSMUST00000062117.14
|
Rap2a
|
RAS related protein 2a |
| chrX_-_73067514 | 1.12 |
ENSMUST00000033769.15
ENSMUST00000114352.8 ENSMUST00000068286.12 ENSMUST00000114360.10 ENSMUST00000114354.10 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr17_+_44263890 | 1.11 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr1_+_75526225 | 1.10 |
ENSMUST00000154101.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr9_+_108782664 | 1.10 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr4_+_152093260 | 1.07 |
ENSMUST00000097773.4
|
Klhl21
|
kelch-like 21 |
| chr4_-_148236103 | 1.05 |
ENSMUST00000132083.2
|
Fbxo6
|
F-box protein 6 |
| chr5_-_108823435 | 1.04 |
ENSMUST00000051757.14
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr7_-_126183716 | 1.03 |
ENSMUST00000150311.8
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr18_-_35631914 | 1.02 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr9_+_7272514 | 1.01 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr9_-_119852624 | 1.00 |
ENSMUST00000111635.4
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr4_-_140376047 | 0.96 |
ENSMUST00000105799.8
ENSMUST00000039204.10 ENSMUST00000097820.9 |
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr1_+_186699613 | 0.95 |
ENSMUST00000045108.2
|
D1Pas1
|
DNA segment, Chr 1, Pasteur Institute 1 |
| chr9_+_108782646 | 0.95 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr11_+_4207557 | 0.94 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr1_+_170060318 | 0.93 |
ENSMUST00000162752.2
|
Sh2d1b2
|
SH2 domain containing 1B2 |
| chr3_-_59102517 | 0.92 |
ENSMUST00000200095.2
|
Gpr87
|
G protein-coupled receptor 87 |
| chr11_-_31621863 | 0.92 |
ENSMUST00000058060.14
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr11_+_94219046 | 0.91 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr3_+_100732768 | 0.91 |
ENSMUST00000054791.9
|
Vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr15_+_80139371 | 0.91 |
ENSMUST00000109605.5
ENSMUST00000229828.2 |
Atf4
|
activating transcription factor 4 |
| chr17_-_25564501 | 0.90 |
ENSMUST00000153118.2
ENSMUST00000146856.3 |
Tpsab1
|
tryptase alpha/beta 1 |
| chr7_+_126376319 | 0.90 |
ENSMUST00000132643.2
|
Ypel3
|
yippee like 3 |
| chr6_-_87327885 | 0.90 |
ENSMUST00000032129.3
|
Gkn1
|
gastrokine 1 |
| chr19_+_5100815 | 0.89 |
ENSMUST00000224178.2
ENSMUST00000225799.3 ENSMUST00000025818.8 |
Rin1
|
Ras and Rab interactor 1 |
| chr11_+_121036969 | 0.87 |
ENSMUST00000039088.9
ENSMUST00000155694.2 |
Tex19.1
|
testis expressed gene 19.1 |
| chrX_-_73067351 | 0.87 |
ENSMUST00000114353.10
ENSMUST00000101458.9 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr9_+_7445822 | 0.87 |
ENSMUST00000034497.8
|
Mmp3
|
matrix metallopeptidase 3 |
| chr11_+_80320558 | 0.86 |
ENSMUST00000173565.2
|
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr18_+_33072194 | 0.85 |
ENSMUST00000042868.6
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr7_+_140659038 | 0.83 |
ENSMUST00000159375.8
|
Pkp3
|
plakophilin 3 |
| chr9_+_37524966 | 0.82 |
ENSMUST00000215474.2
|
Siae
|
sialic acid acetylesterase |
| chr16_-_56537650 | 0.82 |
ENSMUST00000128551.8
|
Tfg
|
Trk-fused gene |
| chr6_+_17463925 | 0.82 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr3_+_87855973 | 0.80 |
ENSMUST00000005019.6
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr1_+_107517726 | 0.80 |
ENSMUST00000000514.11
ENSMUST00000112706.4 |
Serpinb8
|
serine (or cysteine) peptidase inhibitor, clade B, member 8 |
| chr5_+_31274064 | 0.79 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr2_+_160722562 | 0.79 |
ENSMUST00000109456.9
|
Lpin3
|
lipin 3 |
| chr19_+_5100475 | 0.79 |
ENSMUST00000225427.2
|
Rin1
|
Ras and Rab interactor 1 |
| chr11_-_93859064 | 0.79 |
ENSMUST00000107844.3
ENSMUST00000170303.2 |
Nme1
Gm20390
|
NME/NM23 nucleoside diphosphate kinase 1 predicted gene 20390 |
| chr7_-_4781140 | 0.79 |
ENSMUST00000094892.12
|
Il11
|
interleukin 11 |
| chr2_-_25129863 | 0.78 |
ENSMUST00000186719.2
ENSMUST00000043379.5 |
Cysrt1
|
cysteine rich tail 1 |
| chr3_+_79793237 | 0.78 |
ENSMUST00000029567.9
|
Gask1b
|
golgi associated kinase 1B |
| chr5_+_31274046 | 0.77 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr3_-_88243455 | 0.77 |
ENSMUST00000193872.2
|
Tmem79
|
transmembrane protein 79 |
| chr10_-_5872386 | 0.77 |
ENSMUST00000131996.8
ENSMUST00000064225.14 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr8_+_36956345 | 0.77 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr14_-_55995912 | 0.75 |
ENSMUST00000001497.9
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B |
| chr11_-_29464998 | 0.74 |
ENSMUST00000133103.2
ENSMUST00000039900.4 |
Prorsd1
|
prolyl-tRNA synthetase domain containing 1 |
| chr19_+_5497575 | 0.73 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr17_-_31383976 | 0.73 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chrX_+_5959507 | 0.72 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr19_-_11796085 | 0.72 |
ENSMUST00000211047.2
ENSMUST00000075304.14 ENSMUST00000211641.2 |
Stx3
|
syntaxin 3 |
| chr11_+_94218810 | 0.71 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr10_-_5872341 | 0.70 |
ENSMUST00000117676.8
ENSMUST00000019909.8 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr6_+_82029288 | 0.70 |
ENSMUST00000149023.2
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr13_+_93440572 | 0.69 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr8_+_108020132 | 0.69 |
ENSMUST00000151114.8
ENSMUST00000125721.8 ENSMUST00000075922.11 |
Nfat5
|
nuclear factor of activated T cells 5 |
| chr12_+_111504640 | 0.68 |
ENSMUST00000222375.2
ENSMUST00000222388.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr2_+_174602574 | 0.66 |
ENSMUST00000140908.2
|
Edn3
|
endothelin 3 |
| chr18_-_35087355 | 0.66 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr11_-_59927688 | 0.66 |
ENSMUST00000102692.10
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr4_-_141450710 | 0.66 |
ENSMUST00000102484.5
ENSMUST00000177592.2 |
Ddi2
|
DNA-damage inducible protein 2 |
| chr1_-_74932266 | 0.66 |
ENSMUST00000006721.3
|
Cryba2
|
crystallin, beta A2 |
| chr13_+_93440265 | 0.65 |
ENSMUST00000109494.8
|
Homer1
|
homer scaffolding protein 1 |
| chr13_-_8921732 | 0.65 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chrX_-_100103220 | 0.65 |
ENSMUST00000009814.10
|
Tex11
|
testis expressed gene 11 |
| chr19_+_8828132 | 0.65 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr13_+_19362068 | 0.65 |
ENSMUST00000103553.3
|
Trgv7
|
T cell receptor gamma, variable 7 |
| chr7_-_4607040 | 0.64 |
ENSMUST00000166650.3
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr8_+_108020092 | 0.64 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr11_+_73158214 | 0.64 |
ENSMUST00000049676.3
|
Trpv3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr2_+_167263626 | 0.63 |
ENSMUST00000047815.13
ENSMUST00000109218.7 ENSMUST00000073873.5 |
Slc9a8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8 |
| chr1_+_180978491 | 0.63 |
ENSMUST00000134115.8
ENSMUST00000111059.2 |
Cnih4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr9_+_44151962 | 0.62 |
ENSMUST00000092426.5
ENSMUST00000217221.2 ENSMUST00000213891.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr6_+_7555053 | 0.62 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr2_+_103242027 | 0.62 |
ENSMUST00000239273.2
ENSMUST00000164172.8 |
Elf5
|
E74-like factor 5 |
| chr9_+_78522783 | 0.61 |
ENSMUST00000093812.5
|
Cd109
|
CD109 antigen |
| chrX_+_163052367 | 0.61 |
ENSMUST00000145412.8
ENSMUST00000033749.9 |
Pir
|
pirin |
| chr15_-_3333003 | 0.61 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr3_-_92346078 | 0.61 |
ENSMUST00000062160.4
|
Sprr1b
|
small proline-rich protein 1B |
| chr16_+_32219324 | 0.61 |
ENSMUST00000115149.3
|
Tm4sf19
|
transmembrane 4 L six family member 19 |
| chr6_+_17463819 | 0.59 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr13_-_65353757 | 0.58 |
ENSMUST00000223418.2
ENSMUST00000222559.2 ENSMUST00000221659.2 ENSMUST00000222273.2 |
Nlrp4f
|
NLR family, pyrin domain containing 4F |
| chr4_-_45489794 | 0.58 |
ENSMUST00000146236.8
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr17_-_24863956 | 0.58 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr11_-_120358239 | 0.58 |
ENSMUST00000076921.7
|
Arl16
|
ADP-ribosylation factor-like 16 |
| chr15_-_74618533 | 0.58 |
ENSMUST00000057932.8
|
Slurp2
|
secreted Ly6/Plaur domain containing 2 |
| chr15_-_97665524 | 0.57 |
ENSMUST00000128775.9
ENSMUST00000134885.3 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr6_+_17693941 | 0.57 |
ENSMUST00000115420.8
ENSMUST00000115419.8 |
St7
|
suppression of tumorigenicity 7 |
| chr18_+_65183987 | 0.57 |
ENSMUST00000236103.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr17_+_48080113 | 0.57 |
ENSMUST00000160373.8
ENSMUST00000159641.8 |
Tfeb
|
transcription factor EB |
| chr14_-_36690726 | 0.57 |
ENSMUST00000090024.11
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_-_22845124 | 0.57 |
ENSMUST00000115273.10
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr2_+_145627900 | 0.57 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr9_+_32135781 | 0.57 |
ENSMUST00000183121.2
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr7_-_120269462 | 0.56 |
ENSMUST00000127845.2
ENSMUST00000208635.2 ENSMUST00000033178.4 |
Pdzd9
|
PDZ domain containing 9 |
| chr1_-_10790120 | 0.56 |
ENSMUST00000035577.7
|
Cpa6
|
carboxypeptidase A6 |
| chr17_-_43053057 | 0.56 |
ENSMUST00000239223.2
ENSMUST00000113614.3 |
Adgrf2
|
adhesion G protein-coupled receptor F2 |
| chr11_-_121009503 | 0.56 |
ENSMUST00000039146.4
|
Tex19.2
|
testis expressed gene 19.2 |
| chr14_+_79663850 | 0.56 |
ENSMUST00000061222.9
|
Kbtbd7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr11_-_31621727 | 0.55 |
ENSMUST00000109415.2
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr14_-_36857083 | 0.55 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr7_+_30463175 | 0.54 |
ENSMUST00000165887.8
ENSMUST00000085691.11 ENSMUST00000054427.13 ENSMUST00000085688.11 |
Dmkn
|
dermokine |
| chr2_-_17735847 | 0.54 |
ENSMUST00000028080.12
|
Nebl
|
nebulette |
| chrX_-_47763355 | 0.54 |
ENSMUST00000053970.4
|
Gpr119
|
G-protein coupled receptor 119 |
| chr13_+_33187205 | 0.53 |
ENSMUST00000063191.14
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr10_-_43880353 | 0.53 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr9_+_32135540 | 0.53 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr6_-_106725895 | 0.52 |
ENSMUST00000205004.2
|
Il5ra
|
interleukin 5 receptor, alpha |
| chr6_+_124973752 | 0.52 |
ENSMUST00000162000.4
|
Pianp
|
PILR alpha associated neural protein |
| chr15_+_39609320 | 0.52 |
ENSMUST00000227368.2
ENSMUST00000228556.2 ENSMUST00000022913.6 ENSMUST00000228701.2 ENSMUST00000227792.2 |
Dcstamp
|
dendrocyte expressed seven transmembrane protein |
| chr13_+_15638466 | 0.52 |
ENSMUST00000110510.4
|
Gli3
|
GLI-Kruppel family member GLI3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Jun
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 1.4 | 6.8 | GO:1904970 | brush border assembly(GO:1904970) |
| 1.0 | 8.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.9 | 2.8 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.7 | 4.0 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.6 | 3.7 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.6 | 1.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.6 | 1.7 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.5 | 1.6 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.5 | 2.2 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.5 | 1.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.5 | 1.8 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.4 | 2.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.4 | 2.7 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 4.6 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.4 | 3.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 2.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 5.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 6.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.3 | 2.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 1.0 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.3 | 4.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 2.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 1.0 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.2 | 0.9 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 0.9 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 2.5 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 1.1 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.2 | 0.7 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 0.6 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.2 | 2.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 3.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.8 | GO:0042335 | cuticle development(GO:0042335) |
| 0.2 | 3.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 1.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.2 | 0.5 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.2 | 0.5 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.2 | 2.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 2.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 1.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 0.9 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.2 | 3.5 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 2.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.2 | GO:0051036 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.6 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 0.6 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 1.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.5 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.5 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.3 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 0.7 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.3 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.4 | GO:0002767 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.6 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.9 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.4 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.4 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.9 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 1.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:0097184 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 0.1 | 1.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.6 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.1 | 0.2 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.1 | 0.9 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.3 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.3 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 2.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.5 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.5 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.6 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.9 | GO:0045953 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.1 | 0.3 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.8 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.6 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 2.0 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.2 | GO:0071874 | astrocyte activation involved in immune response(GO:0002265) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.6 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 1.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.4 | GO:0060339 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.7 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 2.5 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 6.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 1.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.9 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 3.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.0 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.0 | 1.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.8 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 1.2 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 1.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 15.4 | GO:0043434 | response to peptide hormone(GO:0043434) |
| 0.0 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.7 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.3 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.4 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.6 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 1.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 2.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.4 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.3 | GO:0010572 | positive regulation of platelet activation(GO:0010572) regulation of integrin activation(GO:0033623) |
| 0.0 | 2.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 1.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.9 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.2 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 1.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 2.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.4 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.2 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.1 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 1.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.8 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 10.9 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.8 | 4.6 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.8 | 2.3 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.7 | 3.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.7 | 6.8 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.5 | 1.6 | GO:0044753 | amphisome(GO:0044753) |
| 0.4 | 1.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.3 | 4.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 1.0 | GO:1990794 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.2 | 0.9 | GO:1990589 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 1.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 3.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 0.4 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 5.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 0.8 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 1.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 2.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.6 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 2.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.5 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 1.1 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 3.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 3.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 2.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 3.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 2.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 3.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 2.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 2.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 1.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.1 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 2.2 | 6.7 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.0 | 2.9 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.9 | 3.5 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.7 | 2.7 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.6 | 2.5 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.6 | 3.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.6 | 1.8 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.5 | 1.6 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.4 | 1.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 2.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 1.9 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.4 | 3.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 2.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 1.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 1.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 1.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 5.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 6.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 0.9 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 0.6 | GO:0008127 | quercetin 2,3-dioxygenase activity(GO:0008127) |
| 0.2 | 0.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 1.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 24.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 4.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 2.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 0.7 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.5 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 6.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.3 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 3.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.2 | GO:0052600 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.1 | 0.3 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 2.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.7 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.5 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 8.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 2.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.5 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 1.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 3.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.6 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 1.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.4 | GO:0050542 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 2.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 2.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 2.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 3.4 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 6.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 5.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 2.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 4.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 1.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 1.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 2.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 7.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 5.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 4.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |