Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
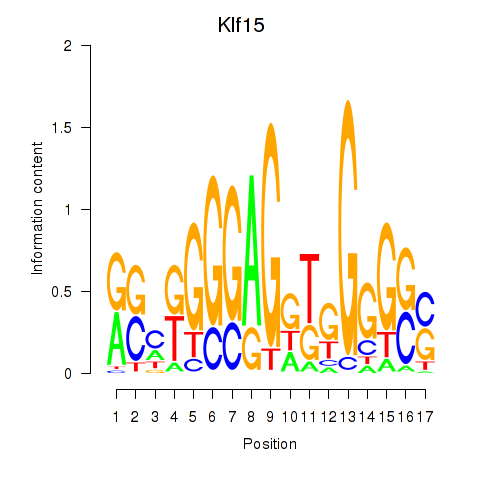
Results for Klf15
Z-value: 1.61
Transcription factors associated with Klf15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf15
|
ENSMUSG00000030087.12 | Kruppel-like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf15 | mm39_v1_chr6_+_90439544_90439565 | 0.79 | 8.8e-09 | Click! |
Activity profile of Klf15 motif
Sorted Z-values of Klf15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_99276310 | 9.43 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr17_+_31605184 | 8.06 |
ENSMUST00000047168.13
ENSMUST00000127929.8 ENSMUST00000134525.9 ENSMUST00000236454.2 ENSMUST00000238091.2 ENSMUST00000235719.2 |
Pde9a
|
phosphodiesterase 9A |
| chr5_+_21391282 | 8.03 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr4_+_139350152 | 7.47 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr8_-_41586713 | 7.37 |
ENSMUST00000155055.2
ENSMUST00000059115.13 ENSMUST00000145860.2 |
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr18_-_38345010 | 7.24 |
ENSMUST00000159405.3
ENSMUST00000160721.8 |
Pcdh1
|
protocadherin 1 |
| chr6_+_54016543 | 7.13 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr2_-_5719302 | 6.59 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr9_-_21838584 | 6.59 |
ENSMUST00000213698.2
|
Tmem205
|
transmembrane protein 205 |
| chr9_-_21838758 | 6.33 |
ENSMUST00000046831.11
ENSMUST00000238930.2 |
Tmem205
|
transmembrane protein 205 |
| chr10_-_53951825 | 6.31 |
ENSMUST00000003843.16
|
Man1a
|
mannosidase 1, alpha |
| chr9_-_43151179 | 6.02 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr11_+_98239230 | 5.87 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr10_-_53951796 | 5.66 |
ENSMUST00000105470.9
|
Man1a
|
mannosidase 1, alpha |
| chr14_-_20844074 | 5.65 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr2_-_147887810 | 5.64 |
ENSMUST00000109964.8
|
Foxa2
|
forkhead box A2 |
| chr16_-_97763780 | 5.61 |
ENSMUST00000232187.2
ENSMUST00000231263.2 ENSMUST00000052089.9 ENSMUST00000063605.15 ENSMUST00000113734.9 ENSMUST00000231560.2 ENSMUST00000232165.2 |
Zbtb21
C2cd2
|
zinc finger and BTB domain containing 21 C2 calcium-dependent domain containing 2 |
| chr7_-_16348862 | 5.28 |
ENSMUST00000171937.2
ENSMUST00000075845.11 |
Arhgap35
|
Rho GTPase activating protein 35 |
| chr9_+_108174052 | 4.40 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chrX_+_169106356 | 4.22 |
ENSMUST00000178693.4
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr4_+_107035566 | 4.18 |
ENSMUST00000030361.11
|
Tmem59
|
transmembrane protein 59 |
| chr17_-_33166346 | 4.16 |
ENSMUST00000139353.8
|
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chr2_+_128660212 | 4.09 |
ENSMUST00000143398.3
ENSMUST00000152210.2 |
Tmem87b
|
transmembrane protein 87B |
| chr9_-_110571645 | 4.09 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr10_-_81262948 | 3.92 |
ENSMUST00000078185.14
ENSMUST00000020461.15 ENSMUST00000105321.10 |
Nfic
|
nuclear factor I/C |
| chr19_+_53891802 | 3.86 |
ENSMUST00000165617.3
|
Pdcd4
|
programmed cell death 4 |
| chr4_+_107035615 | 3.75 |
ENSMUST00000128123.3
ENSMUST00000106753.3 |
Tmem59
|
transmembrane protein 59 |
| chr11_-_100661762 | 3.73 |
ENSMUST00000139341.2
ENSMUST00000017891.14 |
Ghdc
|
GH3 domain containing |
| chr6_-_84564623 | 3.72 |
ENSMUST00000205228.3
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr9_+_21838767 | 3.65 |
ENSMUST00000006403.7
ENSMUST00000170304.9 ENSMUST00000216710.2 |
Ccdc159
|
coiled-coil domain containing 159 |
| chr2_+_32496990 | 3.60 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr14_-_64654397 | 3.47 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr14_-_64654592 | 3.44 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chr4_-_118148471 | 3.34 |
ENSMUST00000222620.2
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr6_+_54017063 | 3.33 |
ENSMUST00000127323.3
|
Chn2
|
chimerin 2 |
| chr1_-_93406515 | 3.26 |
ENSMUST00000170883.8
ENSMUST00000189025.7 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr16_-_95883588 | 3.10 |
ENSMUST00000099502.9
ENSMUST00000023631.17 ENSMUST00000232755.2 |
Brwd1
|
bromodomain and WD repeat domain containing 1 |
| chrX_-_146337046 | 3.10 |
ENSMUST00000112819.9
ENSMUST00000136789.8 |
Lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr4_-_118148537 | 3.08 |
ENSMUST00000049074.13
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr9_+_43996236 | 3.04 |
ENSMUST00000065461.9
ENSMUST00000176416.8 |
Usp2
|
ubiquitin specific peptidase 2 |
| chr10_-_80096793 | 3.00 |
ENSMUST00000020359.7
|
Gamt
|
guanidinoacetate methyltransferase |
| chr11_-_72686627 | 2.97 |
ENSMUST00000079681.6
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr10_-_80096842 | 2.91 |
ENSMUST00000105363.8
|
Gamt
|
guanidinoacetate methyltransferase |
| chr10_+_80192293 | 2.91 |
ENSMUST00000039836.15
ENSMUST00000105351.2 |
Plk5
|
polo like kinase 5 |
| chr17_+_53873964 | 2.79 |
ENSMUST00000000724.15
|
Kat2b
|
K(lysine) acetyltransferase 2B |
| chr5_+_127709302 | 2.73 |
ENSMUST00000118139.3
|
Glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr9_+_107957621 | 2.70 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr5_+_135093056 | 2.65 |
ENSMUST00000071263.7
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr12_+_103498542 | 2.64 |
ENSMUST00000021631.12
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr9_+_107957640 | 2.63 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr1_-_160862364 | 2.63 |
ENSMUST00000177003.2
ENSMUST00000159250.9 ENSMUST00000162226.9 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr14_-_20844034 | 2.62 |
ENSMUST00000226630.2
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr1_-_93406091 | 2.61 |
ENSMUST00000188165.2
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr6_+_54249817 | 2.61 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr9_+_45817795 | 2.58 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr1_-_74924481 | 2.57 |
ENSMUST00000159232.2
ENSMUST00000068631.4 |
Fev
|
FEV transcription factor, ETS family member |
| chr18_-_3337467 | 2.55 |
ENSMUST00000154135.8
ENSMUST00000142690.2 ENSMUST00000025069.11 ENSMUST00000165086.8 ENSMUST00000082141.12 ENSMUST00000149803.8 |
Crem
|
cAMP responsive element modulator |
| chr15_-_89064936 | 2.54 |
ENSMUST00000109331.9
|
Plxnb2
|
plexin B2 |
| chr15_-_78687216 | 2.54 |
ENSMUST00000164826.8
|
Card10
|
caspase recruitment domain family, member 10 |
| chr6_-_113354668 | 2.54 |
ENSMUST00000193384.2
|
Tada3
|
transcriptional adaptor 3 |
| chr15_+_75865604 | 2.53 |
ENSMUST00000089669.6
|
Mapk15
|
mitogen-activated protein kinase 15 |
| chr13_+_51254852 | 2.49 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr2_-_90410922 | 2.49 |
ENSMUST00000168621.3
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr5_-_138980690 | 2.49 |
ENSMUST00000046901.14
ENSMUST00000076095.14 |
Pdgfa
|
platelet derived growth factor, alpha |
| chr12_-_108241597 | 2.44 |
ENSMUST00000222310.2
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr10_-_127502541 | 2.43 |
ENSMUST00000026469.9
|
Nab2
|
Ngfi-A binding protein 2 |
| chr8_+_108020132 | 2.41 |
ENSMUST00000151114.8
ENSMUST00000125721.8 ENSMUST00000075922.11 |
Nfat5
|
nuclear factor of activated T cells 5 |
| chr2_+_128659997 | 2.37 |
ENSMUST00000110325.8
|
Tmem87b
|
transmembrane protein 87B |
| chr15_+_87509413 | 2.36 |
ENSMUST00000068088.8
|
Tafa5
|
TAFA chemokine like family member 5 |
| chrX_+_160500623 | 2.34 |
ENSMUST00000061514.8
|
Rai2
|
retinoic acid induced 2 |
| chr14_-_70757601 | 2.32 |
ENSMUST00000022693.9
|
Bmp1
|
bone morphogenetic protein 1 |
| chr7_+_97229065 | 2.30 |
ENSMUST00000107153.3
|
Rsf1
|
remodeling and spacing factor 1 |
| chr18_-_38068430 | 2.29 |
ENSMUST00000025337.14
|
Diaph1
|
diaphanous related formin 1 |
| chr8_+_108020092 | 2.28 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr4_+_136197066 | 2.27 |
ENSMUST00000170102.8
ENSMUST00000105849.9 ENSMUST00000129230.3 |
Luzp1
|
leucine zipper protein 1 |
| chr1_-_175319842 | 2.24 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr5_-_65650133 | 2.24 |
ENSMUST00000196121.2
|
Gm43552
|
predicted gene 43552 |
| chr13_+_58550499 | 2.20 |
ENSMUST00000225815.2
|
Rmi1
|
RecQ mediated genome instability 1 |
| chr19_-_44017637 | 2.18 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr2_+_140237366 | 2.18 |
ENSMUST00000110061.2
|
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chr2_+_140237229 | 2.14 |
ENSMUST00000110067.8
ENSMUST00000110063.8 ENSMUST00000110064.8 ENSMUST00000110062.8 ENSMUST00000043836.8 ENSMUST00000078027.12 |
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chr7_+_100355798 | 2.14 |
ENSMUST00000107042.9
ENSMUST00000207564.2 ENSMUST00000049053.9 |
Fam168a
|
family with sequence similarity 168, member A |
| chr4_+_131649001 | 2.14 |
ENSMUST00000094666.4
|
Tmem200b
|
transmembrane protein 200B |
| chr10_-_127502467 | 2.12 |
ENSMUST00000099157.4
|
Nab2
|
Ngfi-A binding protein 2 |
| chr18_-_3337679 | 2.11 |
ENSMUST00000150235.8
ENSMUST00000154470.8 |
Crem
|
cAMP responsive element modulator |
| chr4_+_47353277 | 2.09 |
ENSMUST00000044234.14
|
Tgfbr1
|
transforming growth factor, beta receptor I |
| chr2_+_91087668 | 2.08 |
ENSMUST00000111349.9
ENSMUST00000131711.8 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr18_+_35963353 | 2.08 |
ENSMUST00000235169.2
|
Cxxc5
|
CXXC finger 5 |
| chr18_-_38068456 | 2.07 |
ENSMUST00000080033.7
ENSMUST00000115631.8 ENSMUST00000115634.8 |
Diaph1
|
diaphanous related formin 1 |
| chr11_-_115310743 | 2.07 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr15_-_82783978 | 2.05 |
ENSMUST00000230403.2
|
Tcf20
|
transcription factor 20 |
| chr3_-_30563611 | 2.00 |
ENSMUST00000173899.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr7_-_80052491 | 1.99 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr1_-_16589511 | 1.97 |
ENSMUST00000162751.8
ENSMUST00000027052.13 ENSMUST00000149320.9 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr17_+_43978377 | 1.97 |
ENSMUST00000233627.2
ENSMUST00000233437.2 |
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chrX_+_140258381 | 1.93 |
ENSMUST00000112931.8
ENSMUST00000112930.8 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr9_+_60701749 | 1.92 |
ENSMUST00000214354.2
ENSMUST00000050183.7 ENSMUST00000217656.2 |
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr17_+_43978280 | 1.85 |
ENSMUST00000170988.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr7_+_46045862 | 1.84 |
ENSMUST00000025202.8
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr17_+_11059309 | 1.83 |
ENSMUST00000233706.2
|
Prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr19_-_29790352 | 1.78 |
ENSMUST00000099525.5
|
Ranbp6
|
RAN binding protein 6 |
| chr10_-_127098932 | 1.78 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr11_+_115310963 | 1.78 |
ENSMUST00000106533.8
ENSMUST00000123345.2 |
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr4_+_47353217 | 1.78 |
ENSMUST00000007757.15
|
Tgfbr1
|
transforming growth factor, beta receptor I |
| chr13_+_81859460 | 1.74 |
ENSMUST00000057598.7
ENSMUST00000224299.2 |
Mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr10_+_62935430 | 1.74 |
ENSMUST00000044059.5
|
Atoh7
|
atonal bHLH transcription factor 7 |
| chr17_+_86475205 | 1.73 |
ENSMUST00000097275.9
|
Prkce
|
protein kinase C, epsilon |
| chr5_-_124563611 | 1.70 |
ENSMUST00000198420.5
|
Sbno1
|
strawberry notch 1 |
| chr9_-_20657643 | 1.68 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2 |
| chr7_-_24643919 | 1.68 |
ENSMUST00000206705.3
|
Erfl
|
ETS repressor factor like |
| chr2_-_84545504 | 1.66 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr11_-_119438569 | 1.66 |
ENSMUST00000026670.5
|
Nptx1
|
neuronal pentraxin 1 |
| chr1_-_55401851 | 1.63 |
ENSMUST00000114423.7
|
Boll
|
boule homolog, RNA binding protein |
| chr18_+_35962881 | 1.60 |
ENSMUST00000237291.2
|
Cxxc5
|
CXXC finger 5 |
| chr11_-_97464755 | 1.59 |
ENSMUST00000126287.2
ENSMUST00000107590.9 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr17_+_31783708 | 1.59 |
ENSMUST00000097352.11
ENSMUST00000237248.2 ENSMUST00000235869.2 ENSMUST00000175806.9 |
Pknox1
|
Pbx/knotted 1 homeobox |
| chr7_+_100355910 | 1.58 |
ENSMUST00000207875.2
ENSMUST00000208013.2 |
Fam168a
|
family with sequence similarity 168, member A |
| chr18_+_35962832 | 1.57 |
ENSMUST00000060722.8
|
Cxxc5
|
CXXC finger 5 |
| chr2_-_25982160 | 1.55 |
ENSMUST00000114159.9
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr16_-_94657531 | 1.55 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr11_+_115310885 | 1.54 |
ENSMUST00000103035.10
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr9_-_81515865 | 1.53 |
ENSMUST00000183482.2
|
Htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr13_-_9814467 | 1.53 |
ENSMUST00000154994.8
ENSMUST00000146039.2 ENSMUST00000110635.8 ENSMUST00000110638.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr2_+_91087156 | 1.52 |
ENSMUST00000144394.8
ENSMUST00000028694.12 ENSMUST00000168916.8 ENSMUST00000156919.8 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr16_-_95883714 | 1.50 |
ENSMUST00000113827.4
|
Brwd1
|
bromodomain and WD repeat domain containing 1 |
| chr11_-_97464866 | 1.50 |
ENSMUST00000207653.2
ENSMUST00000107593.8 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr15_+_74827488 | 1.49 |
ENSMUST00000191436.7
ENSMUST00000191145.7 |
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr19_+_6547790 | 1.47 |
ENSMUST00000113458.8
ENSMUST00000113459.2 |
Nrxn2
|
neurexin II |
| chr13_-_9814407 | 1.47 |
ENSMUST00000146059.8
ENSMUST00000110637.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr11_+_77821626 | 1.45 |
ENSMUST00000093995.10
ENSMUST00000000646.14 |
Sez6
|
seizure related gene 6 |
| chr17_+_11059248 | 1.44 |
ENSMUST00000191124.7
|
Prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr7_-_28329924 | 1.43 |
ENSMUST00000159095.2
ENSMUST00000159418.8 ENSMUST00000159560.3 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr12_-_98703664 | 1.43 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr13_+_119565669 | 1.42 |
ENSMUST00000173627.8
ENSMUST00000126957.9 ENSMUST00000174691.8 |
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr5_-_124564014 | 1.41 |
ENSMUST00000196329.5
ENSMUST00000196644.5 |
Sbno1
|
strawberry notch 1 |
| chr15_+_99192968 | 1.40 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr5_-_65650269 | 1.39 |
ENSMUST00000121661.8
|
Smim14
|
small integral membrane protein 14 |
| chr7_+_126376319 | 1.35 |
ENSMUST00000132643.2
|
Ypel3
|
yippee like 3 |
| chr3_+_135144287 | 1.35 |
ENSMUST00000196591.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr9_-_45817666 | 1.34 |
ENSMUST00000161187.8
|
Rnf214
|
ring finger protein 214 |
| chr10_-_83173289 | 1.34 |
ENSMUST00000126617.2
|
Slc41a2
|
solute carrier family 41, member 2 |
| chr2_-_120144622 | 1.33 |
ENSMUST00000054651.8
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr13_-_9814901 | 1.31 |
ENSMUST00000223421.2
ENSMUST00000128658.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr5_+_130477642 | 1.30 |
ENSMUST00000111288.4
|
Caln1
|
calneuron 1 |
| chr18_+_77273510 | 1.30 |
ENSMUST00000075290.8
ENSMUST00000079618.11 |
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr12_+_52550775 | 1.29 |
ENSMUST00000219443.2
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr11_-_100650566 | 1.29 |
ENSMUST00000107361.9
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr13_+_58550046 | 1.27 |
ENSMUST00000225828.2
ENSMUST00000042450.10 |
Rmi1
|
RecQ mediated genome instability 1 |
| chr6_+_110622533 | 1.26 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr13_-_46118433 | 1.26 |
ENSMUST00000167708.4
ENSMUST00000091628.11 ENSMUST00000180110.9 |
Atxn1
|
ataxin 1 |
| chr15_-_83609127 | 1.24 |
ENSMUST00000171496.9
ENSMUST00000043634.12 ENSMUST00000076060.12 ENSMUST00000016907.8 |
Scube1
|
signal peptide, CUB domain, EGF-like 1 |
| chr1_+_55445033 | 1.24 |
ENSMUST00000042986.10
|
Plcl1
|
phospholipase C-like 1 |
| chr11_-_116197994 | 1.22 |
ENSMUST00000124281.2
|
Exoc7
|
exocyst complex component 7 |
| chr11_-_3813895 | 1.21 |
ENSMUST00000070552.14
|
Osbp2
|
oxysterol binding protein 2 |
| chr17_-_28569721 | 1.19 |
ENSMUST00000156862.3
|
Tead3
|
TEA domain family member 3 |
| chr4_+_55350043 | 1.19 |
ENSMUST00000030134.9
|
Rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr4_+_97665992 | 1.18 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr12_+_55646209 | 1.18 |
ENSMUST00000051857.5
|
Insm2
|
insulinoma-associated 2 |
| chr17_-_15596230 | 1.16 |
ENSMUST00000014917.8
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr4_+_97665843 | 1.16 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr6_-_113354826 | 1.14 |
ENSMUST00000032410.14
|
Tada3
|
transcriptional adaptor 3 |
| chr1_+_75523166 | 1.14 |
ENSMUST00000138814.2
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr4_+_131648509 | 1.13 |
ENSMUST00000238733.2
|
Tmem200b
|
transmembrane protein 200B |
| chr5_-_124563969 | 1.12 |
ENSMUST00000065263.12
|
Sbno1
|
strawberry notch 1 |
| chr7_+_127111576 | 1.12 |
ENSMUST00000186672.7
|
Srcap
|
Snf2-related CREBBP activator protein |
| chr4_+_129878627 | 1.12 |
ENSMUST00000120204.8
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr16_-_13977084 | 1.11 |
ENSMUST00000090300.6
|
Marf1
|
meiosis regulator and mRNA stability 1 |
| chr2_-_63014622 | 1.10 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr5_+_30745447 | 1.09 |
ENSMUST00000066295.5
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr10_-_77879414 | 1.05 |
ENSMUST00000145975.8
ENSMUST00000130972.8 ENSMUST00000128241.8 ENSMUST00000155021.8 ENSMUST00000140636.8 ENSMUST00000148469.8 ENSMUST00000019257.15 ENSMUST00000105395.9 ENSMUST00000156417.8 ENSMUST00000105396.9 ENSMUST00000154374.2 |
Aire
|
autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) |
| chr7_-_28330322 | 1.05 |
ENSMUST00000040112.5
ENSMUST00000239470.2 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr18_-_89787603 | 1.05 |
ENSMUST00000097495.5
|
Dok6
|
docking protein 6 |
| chr7_+_16609227 | 1.03 |
ENSMUST00000108493.3
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr11_+_76563281 | 1.02 |
ENSMUST00000056184.2
|
Bhlha9
|
basic helix-loop-helix family, member a9 |
| chr17_-_25652236 | 1.01 |
ENSMUST00000159623.2
|
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr1_+_105918383 | 1.01 |
ENSMUST00000119166.8
|
Zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr17_+_27152012 | 1.00 |
ENSMUST00000073724.7
ENSMUST00000237189.2 |
Phf1
|
PHD finger protein 1 |
| chr2_-_26335187 | 1.00 |
ENSMUST00000114082.9
ENSMUST00000091252.5 |
Sec16a
|
SEC16 homolog A, endoplasmic reticulum export factor |
| chr5_-_135092904 | 0.98 |
ENSMUST00000111205.8
ENSMUST00000141309.2 |
Bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr19_+_48194464 | 0.98 |
ENSMUST00000078880.6
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr10_-_84938350 | 0.97 |
ENSMUST00000059383.8
ENSMUST00000216889.2 |
Fhl4
|
four and a half LIM domains 4 |
| chr8_-_35432783 | 0.96 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chrX_-_58179754 | 0.95 |
ENSMUST00000033473.12
|
Fgf13
|
fibroblast growth factor 13 |
| chr9_+_21077010 | 0.95 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr1_+_93406809 | 0.94 |
ENSMUST00000112912.8
|
Septin2
|
septin 2 |
| chr1_+_105918114 | 0.94 |
ENSMUST00000118196.8
|
Zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr11_-_52173391 | 0.94 |
ENSMUST00000086844.10
|
Tcf7
|
transcription factor 7, T cell specific |
| chr11_-_116197523 | 0.94 |
ENSMUST00000133468.2
ENSMUST00000106411.10 ENSMUST00000106413.10 ENSMUST00000021147.14 |
Exoc7
|
exocyst complex component 7 |
| chr18_+_61178211 | 0.92 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr5_-_71815318 | 0.92 |
ENSMUST00000199357.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr1_-_16589425 | 0.90 |
ENSMUST00000159558.8
ENSMUST00000054668.13 ENSMUST00000162627.8 ENSMUST00000162007.8 ENSMUST00000128957.9 ENSMUST00000115359.10 ENSMUST00000151888.8 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr4_+_155334417 | 0.89 |
ENSMUST00000178473.8
ENSMUST00000105627.8 ENSMUST00000097747.9 |
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr3_+_94385661 | 0.87 |
ENSMUST00000200342.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr2_-_63014514 | 0.86 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr13_-_99653045 | 0.86 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr17_-_25716189 | 0.86 |
ENSMUST00000165183.10
ENSMUST00000051864.5 |
Sstr5
|
somatostatin receptor 5 |
| chr13_-_58261406 | 0.85 |
ENSMUST00000160860.9
|
Klhl3
|
kelch-like 3 |
| chr11_-_100541519 | 0.85 |
ENSMUST00000103119.10
|
Zfp385c
|
zinc finger protein 385C |
| chr13_-_9814979 | 0.84 |
ENSMUST00000110634.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr9_+_89791943 | 0.84 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
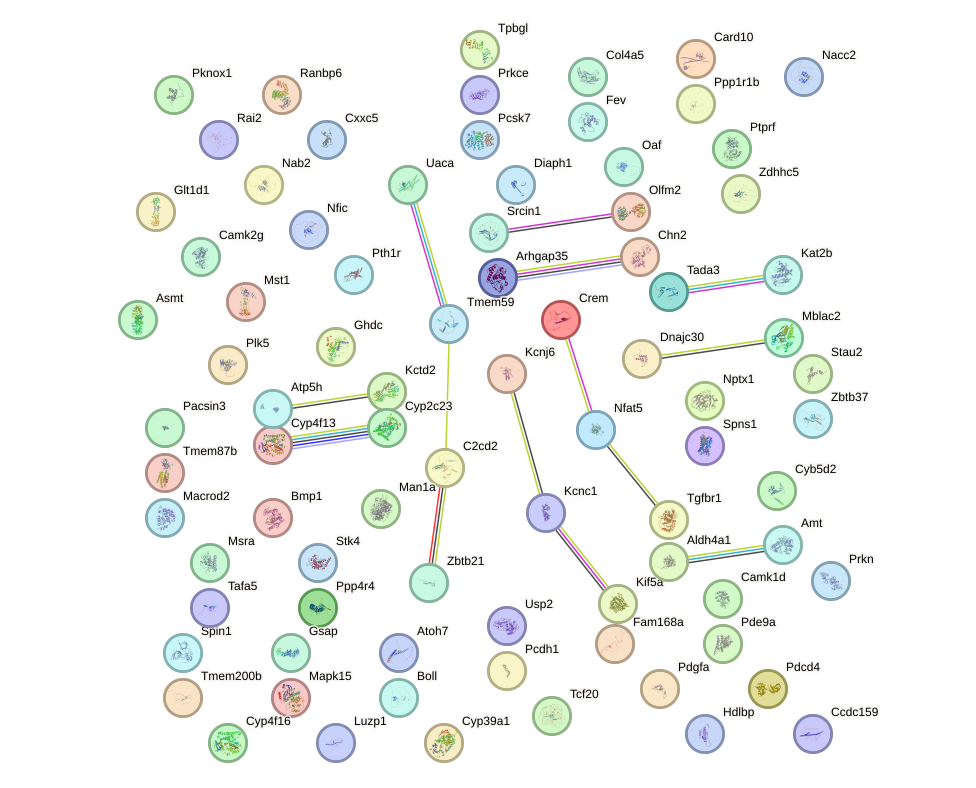
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 2.5 | 7.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.0 | 5.9 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 1.9 | 5.6 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.5 | 4.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.4 | 4.2 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 1.3 | 3.9 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 1.3 | 3.9 | GO:0060938 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) negative regulation of myofibroblast differentiation(GO:1904761) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 1.3 | 3.8 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 1.2 | 2.5 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 1.2 | 5.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.1 | 6.4 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.9 | 3.7 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.9 | 0.9 | GO:2000591 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.8 | 5.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.7 | 2.9 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.7 | 6.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.7 | 2.6 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.6 | 3.7 | GO:0035981 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.6 | 4.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.6 | 5.3 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.6 | 3.5 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.6 | 2.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.5 | 1.6 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.5 | 4.6 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.5 | 2.0 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.4 | 1.3 | GO:0071603 | trabecular meshwork development(GO:0002930) endothelial cell-cell adhesion(GO:0071603) |
| 0.4 | 8.0 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.4 | 3.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 12.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.4 | 1.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.4 | 1.2 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) |
| 0.4 | 1.9 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.4 | 8.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 1.8 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.4 | 2.8 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.3 | 1.7 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 8.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.3 | 1.0 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.3 | 1.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.3 | 4.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 1.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.3 | 1.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.3 | 2.6 | GO:0051611 | regulation of serotonin uptake(GO:0051611) |
| 0.3 | 0.8 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.3 | 1.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.3 | 1.1 | GO:2000410 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) regulation of thymocyte migration(GO:2000410) |
| 0.3 | 9.9 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 0.7 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 4.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 0.7 | GO:1902868 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 1.7 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.2 | 0.7 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.2 | 8.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.6 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.2 | 1.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 0.8 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.2 | 2.9 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 2.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 1.3 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.2 | 0.5 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 2.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 1.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.9 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 3.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 2.3 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 1.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 4.2 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.1 | 0.9 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 2.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 4.8 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.1 | 0.6 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.7 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 1.5 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.1 | 0.7 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 4.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 5.5 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.1 | 2.0 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.1 | 0.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.3 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.8 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 0.9 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 1.9 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 2.9 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 3.6 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 0.4 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.5 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 1.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 4.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 0.5 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 1.0 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 1.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 3.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 2.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.3 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.1 | 1.5 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.1 | 0.4 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.6 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 1.3 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.1 | 1.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.9 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.1 | 0.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 1.4 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 2.8 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 1.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.8 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 3.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.0 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 1.5 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.7 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 1.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 1.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 5.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:1904628 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 1.4 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 1.4 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 2.6 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.8 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.4 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 1.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 1.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.4 | 1.9 | GO:0043293 | apoptosome(GO:0043293) |
| 0.3 | 6.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 1.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.3 | 1.9 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 2.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 0.7 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.1 | 12.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 2.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 2.0 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 5.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 2.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 11.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 2.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 4.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 4.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.8 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 3.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.8 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 14.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 5.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 2.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 3.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 4.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.0 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 1.7 | 6.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 1.6 | 6.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.3 | 3.8 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.9 | 12.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.6 | 3.7 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.6 | 4.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.4 | 1.7 | GO:0035276 | calcium-independent protein kinase C activity(GO:0004699) ethanol binding(GO:0035276) |
| 0.4 | 3.9 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.4 | 3.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 1.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 1.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.3 | 0.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 14.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 8.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 3.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 7.5 | GO:0016646 | aldehyde dehydrogenase (NAD) activity(GO:0004029) oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.3 | 3.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 0.7 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.2 | 5.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 1.0 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.2 | 2.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.7 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 2.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 2.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 3.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 3.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 4.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 2.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 2.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 2.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 8.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 6.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 5.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.3 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 2.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.4 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 3.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.3 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 3.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.0 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 5.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.7 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 3.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 2.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.1 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 2.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 3.0 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 4.4 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 9.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 1.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 5.6 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) sulfate binding(GO:0043199) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 4.8 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 3.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 9.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 10.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 4.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 6.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 6.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 5.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 2.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 4.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 3.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 8.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 7.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 3.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 1.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 5.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 5.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.3 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.1 | 17.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.7 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 1.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 12.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |