Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
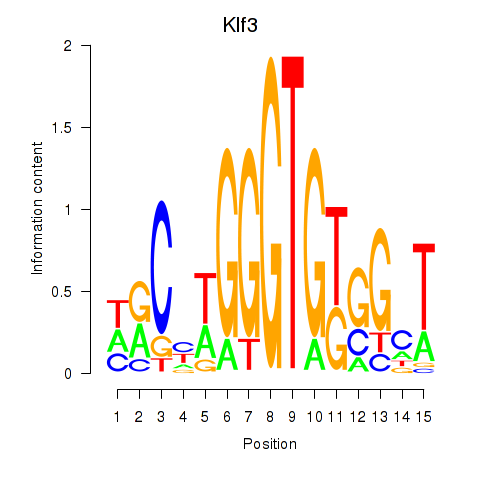
Results for Klf3
Z-value: 0.31
Transcription factors associated with Klf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf3
|
ENSMUSG00000029178.15 | Kruppel-like factor 3 (basic) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf3 | mm39_v1_chr5_+_64969679_64969727 | 0.31 | 6.9e-02 | Click! |
Activity profile of Klf3 motif
Sorted Z-values of Klf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_101295787 | 2.74 |
ENSMUST00000050551.10
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chrX_-_101295458 | 2.48 |
ENSMUST00000101336.10
ENSMUST00000136277.2 |
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr7_-_103477126 | 1.39 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr15_+_78810919 | 1.39 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr19_+_10019023 | 1.31 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr7_+_24069680 | 1.26 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr19_+_10018914 | 0.85 |
ENSMUST00000115995.4
|
Fads3
|
fatty acid desaturase 3 |
| chr5_+_137628377 | 0.40 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chrX_-_73290140 | 0.37 |
ENSMUST00000101454.9
ENSMUST00000033699.13 |
Flna
|
filamin, alpha |
| chr7_-_140676623 | 0.34 |
ENSMUST00000209352.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chrX_-_73289970 | 0.29 |
ENSMUST00000130007.8
|
Flna
|
filamin, alpha |
| chr7_-_105218472 | 0.28 |
ENSMUST00000187683.7
ENSMUST00000210079.2 ENSMUST00000187051.7 ENSMUST00000189265.7 ENSMUST00000190369.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr7_-_142628298 | 0.26 |
ENSMUST00000148715.8
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr7_-_105217851 | 0.25 |
ENSMUST00000188368.7
ENSMUST00000187057.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr13_+_55468313 | 0.20 |
ENSMUST00000021942.8
|
Prelid1
|
PRELI domain containing 1 |
| chr15_-_102630496 | 0.19 |
ENSMUST00000171838.2
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr15_-_102630589 | 0.17 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr12_+_33365371 | 0.16 |
ENSMUST00000154742.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chrX_-_50031587 | 0.16 |
ENSMUST00000060650.7
|
Frmd7
|
FERM domain containing 7 |
| chrX_+_7750558 | 0.15 |
ENSMUST00000208640.2
ENSMUST00000207114.2 ENSMUST00000208633.2 ENSMUST00000208397.2 ENSMUST00000153620.3 ENSMUST00000123277.8 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr19_-_5738177 | 0.14 |
ENSMUST00000068169.12
|
Pcnx3
|
pecanex homolog 3 |
| chr13_+_89687915 | 0.13 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr19_-_53378990 | 0.12 |
ENSMUST00000025997.7
|
Smndc1
|
survival motor neuron domain containing 1 |
| chr3_-_108306267 | 0.12 |
ENSMUST00000147251.2
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr7_-_140676596 | 0.12 |
ENSMUST00000209199.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr12_+_87073338 | 0.11 |
ENSMUST00000110187.8
ENSMUST00000156162.8 |
Tmem63c
|
transmembrane protein 63c |
| chr3_+_89173859 | 0.10 |
ENSMUST00000040824.2
|
Dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr7_-_142627774 | 0.09 |
ENSMUST00000136602.8
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr2_+_119863909 | 0.09 |
ENSMUST00000126150.2
|
Pla2g4b
|
phospholipase A2, group IVB (cytosolic) |
| chr3_-_88241735 | 0.08 |
ENSMUST00000107552.2
|
Tmem79
|
transmembrane protein 79 |
| chr14_+_63284438 | 0.07 |
ENSMUST00000067990.8
ENSMUST00000111203.2 |
Defb42
|
defensin beta 42 |
| chr3_+_89986831 | 0.06 |
ENSMUST00000029549.16
|
Tpm3
|
tropomyosin 3, gamma |
| chr1_+_85992341 | 0.05 |
ENSMUST00000027432.9
|
Psmd1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr17_-_23941757 | 0.03 |
ENSMUST00000178006.8
|
1520401A03Rik
|
RIKEN cDNA 1520401A03 gene |
| chr3_+_89986925 | 0.02 |
ENSMUST00000118566.8
ENSMUST00000119158.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr4_-_41774097 | 0.02 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr14_+_123897383 | 0.02 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.5 | 1.4 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.2 | 0.7 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.5 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 1.3 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.2 | GO:1901857 | negative regulation of mitochondrial membrane potential(GO:0010917) positive regulation of cellular respiration(GO:1901857) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.7 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.5 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031720 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.3 | 1.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 5.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 0.7 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 1.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |