Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
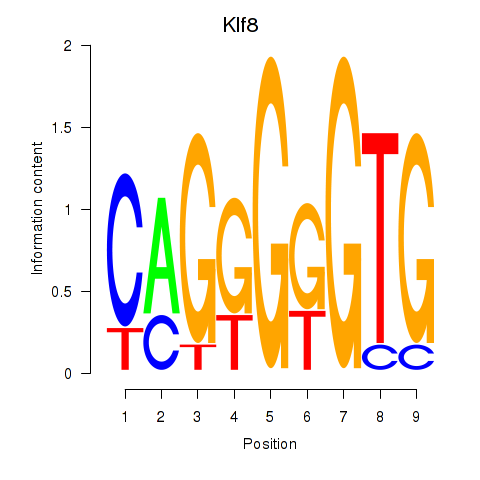
Results for Klf8
Z-value: 0.87
Transcription factors associated with Klf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf8
|
ENSMUSG00000041649.14 | Kruppel-like factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf8 | mm39_v1_chrX_+_152020744_152020838 | -0.17 | 3.2e-01 | Click! |
Activity profile of Klf8 motif
Sorted Z-values of Klf8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_83637766 | 3.39 |
ENSMUST00000070832.3
|
Wfdc21
|
WAP four-disulfide core domain 21 |
| chr19_-_6117815 | 1.76 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr10_+_79716876 | 1.73 |
ENSMUST00000166201.2
|
Prtn3
|
proteinase 3 |
| chrX_-_7834057 | 1.72 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr7_-_30623592 | 1.48 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr7_-_126625657 | 1.44 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr7_-_126625617 | 1.21 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_+_32236945 | 1.16 |
ENSMUST00000101387.4
|
Hbq1b
|
hemoglobin, theta 1B |
| chr2_-_32271833 | 1.13 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr7_-_127529238 | 1.07 |
ENSMUST00000032988.10
ENSMUST00000206124.2 |
Prss8
|
protease, serine 8 (prostasin) |
| chr7_-_28947882 | 1.06 |
ENSMUST00000032808.6
|
2200002D01Rik
|
RIKEN cDNA 2200002D01 gene |
| chr11_-_5900019 | 1.06 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr7_+_24584076 | 1.05 |
ENSMUST00000153451.9
ENSMUST00000108429.8 |
Rps19
|
ribosomal protein S19 |
| chr10_+_75399920 | 1.00 |
ENSMUST00000141062.8
ENSMUST00000152657.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr19_+_6952580 | 0.95 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr14_-_30850795 | 0.94 |
ENSMUST00000049732.11
ENSMUST00000090205.5 ENSMUST00000064032.10 |
Smim4
|
small integral membrane protein 4 |
| chr17_+_29487881 | 0.92 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr6_-_72935171 | 0.91 |
ENSMUST00000114049.2
|
Tmsb10
|
thymosin, beta 10 |
| chr19_+_6952319 | 0.91 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr7_-_27055405 | 0.90 |
ENSMUST00000003857.7
|
Shkbp1
|
Sh3kbp1 binding protein 1 |
| chr15_+_78810919 | 0.88 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr9_-_21202353 | 0.87 |
ENSMUST00000086374.8
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr7_-_103477126 | 0.86 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr9_-_21202545 | 0.85 |
ENSMUST00000215619.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr4_-_43040278 | 0.84 |
ENSMUST00000107958.8
ENSMUST00000107959.8 ENSMUST00000152846.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr8_-_105991219 | 0.83 |
ENSMUST00000034359.10
|
Tradd
|
TNFRSF1A-associated via death domain |
| chr2_-_29983618 | 0.83 |
ENSMUST00000081838.7
ENSMUST00000102865.11 |
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr14_-_70864448 | 0.82 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr7_+_24583994 | 0.82 |
ENSMUST00000108428.8
|
Rps19
|
ribosomal protein S19 |
| chr17_-_57554631 | 0.80 |
ENSMUST00000233568.2
ENSMUST00000005975.8 |
Gpr108
|
G protein-coupled receptor 108 |
| chr8_+_23464860 | 0.79 |
ENSMUST00000110688.9
ENSMUST00000121802.9 |
Ank1
|
ankyrin 1, erythroid |
| chr17_+_29487762 | 0.79 |
ENSMUST00000064709.13
ENSMUST00000234711.2 |
BC004004
|
cDNA sequence BC004004 |
| chr6_+_124908389 | 0.77 |
ENSMUST00000180095.4
|
Mlf2
|
myeloid leukemia factor 2 |
| chr19_+_5540483 | 0.77 |
ENSMUST00000209469.2
ENSMUST00000116560.3 |
Cfl1
|
cofilin 1, non-muscle |
| chr2_-_164197987 | 0.76 |
ENSMUST00000165980.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr7_+_126294527 | 0.76 |
ENSMUST00000130498.2
|
Bola2
|
bolA-like 2 (E. coli) |
| chr10_-_80426125 | 0.74 |
ENSMUST00000187646.2
ENSMUST00000191440.7 ENSMUST00000003436.12 |
Abhd17a
|
abhydrolase domain containing 17A |
| chr12_+_109418759 | 0.71 |
ENSMUST00000056110.15
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr17_-_24915121 | 0.70 |
ENSMUST00000046839.10
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr2_-_164198427 | 0.70 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr7_+_28988724 | 0.68 |
ENSMUST00000207714.2
ENSMUST00000048187.6 |
Ppp1r14a
|
protein phosphatase 1, regulatory inhibitor subunit 14A |
| chr18_-_34505544 | 0.67 |
ENSMUST00000236887.2
|
Reep5
|
receptor accessory protein 5 |
| chr15_+_78290896 | 0.67 |
ENSMUST00000167140.8
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr5_+_31205971 | 0.66 |
ENSMUST00000013766.13
ENSMUST00000201773.4 ENSMUST00000200748.4 ENSMUST00000201136.2 |
Atraid
|
all-trans retinoic acid induced differentiation factor |
| chr16_+_17884267 | 0.65 |
ENSMUST00000151266.8
ENSMUST00000066027.14 ENSMUST00000155387.8 |
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr9_-_21202693 | 0.64 |
ENSMUST00000213407.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr17_-_34219225 | 0.64 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr19_+_5540591 | 0.63 |
ENSMUST00000237122.2
|
Cfl1
|
cofilin 1, non-muscle |
| chr2_-_38534099 | 0.62 |
ENSMUST00000028083.6
|
Psmb7
|
proteasome (prosome, macropain) subunit, beta type 7 |
| chr8_+_105991280 | 0.61 |
ENSMUST00000036221.12
|
Fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr17_+_35117905 | 0.61 |
ENSMUST00000097342.10
ENSMUST00000013931.12 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr9_-_114610879 | 0.61 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr6_+_124908439 | 0.61 |
ENSMUST00000032214.14
|
Mlf2
|
myeloid leukemia factor 2 |
| chrX_-_72965524 | 0.60 |
ENSMUST00000114389.10
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr1_+_135945705 | 0.59 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr7_-_45750050 | 0.58 |
ENSMUST00000209291.2
|
Kcnj11
|
potassium inwardly rectifying channel, subfamily J, member 11 |
| chr1_+_135746330 | 0.57 |
ENSMUST00000038760.10
|
Lad1
|
ladinin |
| chr15_+_78290975 | 0.57 |
ENSMUST00000043865.8
ENSMUST00000231159.2 ENSMUST00000169133.8 |
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr17_-_46956920 | 0.56 |
ENSMUST00000233974.2
|
Klc4
|
kinesin light chain 4 |
| chrX_-_72965434 | 0.55 |
ENSMUST00000096316.4
ENSMUST00000114390.8 ENSMUST00000114391.10 ENSMUST00000114387.8 |
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr1_+_135945798 | 0.53 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr13_+_120151982 | 0.52 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr15_+_100768551 | 0.52 |
ENSMUST00000082209.13
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr13_+_73476629 | 0.51 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr17_-_26014613 | 0.50 |
ENSMUST00000235889.2
|
Gm50367
|
predicted gene, 50367 |
| chrX_-_72965536 | 0.50 |
ENSMUST00000033763.15
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr17_+_35117438 | 0.50 |
ENSMUST00000114033.9
ENSMUST00000078061.13 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr2_+_143757193 | 0.49 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr4_+_127062924 | 0.48 |
ENSMUST00000046659.14
|
Dlgap3
|
DLG associated protein 3 |
| chr5_+_115373895 | 0.47 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr7_+_28937746 | 0.47 |
ENSMUST00000108238.8
ENSMUST00000032809.10 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr11_-_88608958 | 0.47 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr12_-_118265163 | 0.46 |
ENSMUST00000221844.2
|
Sp4
|
trans-acting transcription factor 4 |
| chr11_+_120489358 | 0.45 |
ENSMUST00000093140.5
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr2_+_122479770 | 0.45 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr19_+_6547790 | 0.45 |
ENSMUST00000113458.8
ENSMUST00000113459.2 |
Nrxn2
|
neurexin II |
| chr13_+_108452930 | 0.44 |
ENSMUST00000171178.2
|
Depdc1b
|
DEP domain containing 1B |
| chr14_-_30850881 | 0.44 |
ENSMUST00000203261.3
|
Smim4
|
small integral membrane protein 4 |
| chr17_+_46957151 | 0.44 |
ENSMUST00000002844.14
ENSMUST00000113429.8 ENSMUST00000113430.2 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr2_-_90735171 | 0.43 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr14_-_55828511 | 0.43 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr5_+_31079177 | 0.42 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chrX_+_93278203 | 0.41 |
ENSMUST00000153900.8
|
Klhl15
|
kelch-like 15 |
| chr15_-_75439013 | 0.41 |
ENSMUST00000156032.2
ENSMUST00000127095.8 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr11_+_70548022 | 0.41 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr11_-_115310743 | 0.41 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr9_+_114807546 | 0.41 |
ENSMUST00000183104.8
|
Osbpl10
|
oxysterol binding protein-like 10 |
| chr15_-_89258034 | 0.40 |
ENSMUST00000228977.2
|
Sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr19_+_6919691 | 0.40 |
ENSMUST00000113426.8
ENSMUST00000238184.2 ENSMUST00000113423.10 ENSMUST00000237334.2 ENSMUST00000235240.2 |
Bad
|
BCL2-associated agonist of cell death |
| chrX_+_74460275 | 0.39 |
ENSMUST00000118428.8
ENSMUST00000114074.8 ENSMUST00000133781.8 |
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr2_-_122199604 | 0.39 |
ENSMUST00000151130.8
|
Shf
|
Src homology 2 domain containing F |
| chr15_-_89258012 | 0.39 |
ENSMUST00000167643.4
|
Sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr1_+_172327569 | 0.39 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr7_+_64151838 | 0.38 |
ENSMUST00000205604.2
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr5_+_31078911 | 0.38 |
ENSMUST00000201571.4
|
Khk
|
ketohexokinase |
| chr4_+_109264284 | 0.38 |
ENSMUST00000064129.14
ENSMUST00000106619.8 |
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr8_+_73449907 | 0.38 |
ENSMUST00000109950.5
ENSMUST00000004494.16 ENSMUST00000212095.2 ENSMUST00000212096.2 |
Sin3b
|
transcriptional regulator, SIN3B (yeast) |
| chr3_+_90448453 | 0.38 |
ENSMUST00000107333.8
ENSMUST00000107331.8 ENSMUST00000098910.3 |
S100a16
|
S100 calcium binding protein A16 |
| chr11_-_69838971 | 0.38 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr17_-_71158703 | 0.38 |
ENSMUST00000166395.9
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr9_+_108765701 | 0.37 |
ENSMUST00000026743.14
ENSMUST00000194047.3 |
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr5_+_139777263 | 0.37 |
ENSMUST00000018287.10
|
Mafk
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
| chr12_-_69274936 | 0.37 |
ENSMUST00000221411.2
ENSMUST00000021359.7 |
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr5_+_32616566 | 0.36 |
ENSMUST00000202078.2
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr7_-_30754792 | 0.36 |
ENSMUST00000206328.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_30755007 | 0.36 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr16_-_18630365 | 0.35 |
ENSMUST00000096990.10
|
Cdc45
|
cell division cycle 45 |
| chr7_-_3551003 | 0.34 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr1_+_190830522 | 0.34 |
ENSMUST00000027943.6
|
Batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr7_-_19133783 | 0.34 |
ENSMUST00000047170.10
ENSMUST00000108459.9 |
Klc3
|
kinesin light chain 3 |
| chr4_-_150998857 | 0.34 |
ENSMUST00000105675.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr12_-_103597663 | 0.34 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr7_+_28151370 | 0.33 |
ENSMUST00000190954.7
|
Lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr8_+_94537910 | 0.33 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr5_-_123270449 | 0.33 |
ENSMUST00000186469.7
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr13_-_43324824 | 0.32 |
ENSMUST00000220787.2
|
Tbc1d7
|
TBC1 domain family, member 7 |
| chr5_+_129924564 | 0.32 |
ENSMUST00000041466.14
|
Zbed5
|
zinc finger, BED type containing 5 |
| chr2_+_156681991 | 0.31 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr8_-_64659004 | 0.31 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chrX_-_72703330 | 0.31 |
ENSMUST00000114473.8
ENSMUST00000002087.14 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chrX_-_50294867 | 0.30 |
ENSMUST00000114876.9
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr12_+_71063431 | 0.30 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chrX_+_74460234 | 0.30 |
ENSMUST00000033544.14
|
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr14_+_54713557 | 0.30 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr11_-_62539284 | 0.30 |
ENSMUST00000057194.9
|
Lrrc75a
|
leucine rich repeat containing 75A |
| chr5_-_123270702 | 0.30 |
ENSMUST00000031401.6
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr11_-_67812960 | 0.29 |
ENSMUST00000021288.10
ENSMUST00000108677.2 |
Usp43
|
ubiquitin specific peptidase 43 |
| chr11_-_88609048 | 0.29 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr18_+_62086122 | 0.29 |
ENSMUST00000051720.6
ENSMUST00000235860.2 |
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr4_-_128699838 | 0.29 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chr11_-_102837514 | 0.28 |
ENSMUST00000057849.6
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr15_+_79999643 | 0.28 |
ENSMUST00000135727.2
|
Syngr1
|
synaptogyrin 1 |
| chr9_-_45847344 | 0.28 |
ENSMUST00000034590.4
|
Tagln
|
transgelin |
| chr11_+_72889889 | 0.28 |
ENSMUST00000021141.14
|
P2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr16_+_93480061 | 0.28 |
ENSMUST00000039620.7
|
Cbr3
|
carbonyl reductase 3 |
| chr17_+_23879448 | 0.27 |
ENSMUST00000062967.10
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr9_-_44199626 | 0.27 |
ENSMUST00000161408.2
|
Abcg4
|
ATP binding cassette subfamily G member 4 |
| chrX_+_7750483 | 0.26 |
ENSMUST00000115663.10
ENSMUST00000096514.11 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr1_-_106641940 | 0.26 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr9_-_31824758 | 0.26 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chr8_+_94537460 | 0.26 |
ENSMUST00000034198.15
ENSMUST00000125716.8 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr2_-_52566583 | 0.25 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr2_+_156681927 | 0.25 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr4_-_155127601 | 0.25 |
ENSMUST00000135665.9
|
Plch2
|
phospholipase C, eta 2 |
| chr6_-_136150491 | 0.25 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr7_+_28937898 | 0.25 |
ENSMUST00000138128.3
ENSMUST00000142519.3 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr11_-_120538928 | 0.25 |
ENSMUST00000239158.2
ENSMUST00000026134.3 |
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr11_-_86574586 | 0.25 |
ENSMUST00000018315.10
|
Vmp1
|
vacuole membrane protein 1 |
| chrX_+_7439839 | 0.25 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr8_+_86026318 | 0.24 |
ENSMUST00000170141.3
ENSMUST00000034132.13 |
Orc6
|
origin recognition complex, subunit 6 |
| chr4_+_129384983 | 0.24 |
ENSMUST00000106043.3
|
Fam229a
|
family with sequence similarity 229, member A |
| chr2_-_168583670 | 0.24 |
ENSMUST00000029060.11
|
Atp9a
|
ATPase, class II, type 9A |
| chr10_-_90959853 | 0.24 |
ENSMUST00000170810.8
ENSMUST00000076694.13 |
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr13_-_99584091 | 0.24 |
ENSMUST00000223725.2
|
Map1b
|
microtubule-associated protein 1B |
| chr4_-_130068902 | 0.24 |
ENSMUST00000105998.8
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chrX_+_73298342 | 0.23 |
ENSMUST00000096424.11
|
Emd
|
emerin |
| chr18_+_84738144 | 0.23 |
ENSMUST00000161429.3
ENSMUST00000052501.8 |
Dipk1c
|
divergent protein kinase domain 1C |
| chr8_+_114932312 | 0.23 |
ENSMUST00000049509.7
ENSMUST00000150963.2 |
Vat1l
|
vesicle amine transport protein 1 like |
| chr11_+_70548513 | 0.23 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr9_-_44199428 | 0.23 |
ENSMUST00000160384.2
|
Abcg4
|
ATP binding cassette subfamily G member 4 |
| chr5_+_122347792 | 0.22 |
ENSMUST00000072602.14
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr7_+_25005510 | 0.22 |
ENSMUST00000119703.8
ENSMUST00000205639.3 ENSMUST00000108409.2 |
Tmem145
|
transmembrane protein 145 |
| chr12_-_79343040 | 0.22 |
ENSMUST00000218377.2
ENSMUST00000021547.8 |
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr10_-_90959817 | 0.22 |
ENSMUST00000164505.2
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr14_-_47025724 | 0.21 |
ENSMUST00000146629.3
ENSMUST00000015903.12 |
Cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr6_-_52185674 | 0.21 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr11_-_101357046 | 0.21 |
ENSMUST00000040430.8
|
Vat1
|
vesicle amine transport 1 |
| chr7_-_136915602 | 0.21 |
ENSMUST00000210774.2
|
Ebf3
|
early B cell factor 3 |
| chr3_+_129007599 | 0.21 |
ENSMUST00000042587.12
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr6_+_124908341 | 0.21 |
ENSMUST00000203021.3
|
Mlf2
|
myeloid leukemia factor 2 |
| chr19_-_5962798 | 0.21 |
ENSMUST00000118623.2
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr2_-_26096547 | 0.21 |
ENSMUST00000028302.8
|
Lhx3
|
LIM homeobox protein 3 |
| chr11_-_45835737 | 0.20 |
ENSMUST00000129820.8
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11 |
| chr11_+_69945157 | 0.20 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr7_+_64151435 | 0.20 |
ENSMUST00000032732.15
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr13_+_55517545 | 0.20 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chrX_-_50294652 | 0.20 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr5_+_147206769 | 0.20 |
ENSMUST00000085591.7
|
Pdx1
|
pancreatic and duodenal homeobox 1 |
| chr3_+_96543143 | 0.20 |
ENSMUST00000165842.3
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr17_+_25992761 | 0.20 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr17_-_37523969 | 0.20 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr16_+_20448547 | 0.19 |
ENSMUST00000003898.12
ENSMUST00000122306.8 ENSMUST00000133344.8 |
Ece2
|
endothelin converting enzyme 2 |
| chr19_+_45991907 | 0.19 |
ENSMUST00000099393.4
|
Hps6
|
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| chr13_-_114595122 | 0.19 |
ENSMUST00000231252.2
|
Fst
|
follistatin |
| chr2_-_91540864 | 0.19 |
ENSMUST00000028678.9
ENSMUST00000076803.12 |
Atg13
|
autophagy related 13 |
| chr8_+_107237483 | 0.19 |
ENSMUST00000080797.8
|
Cdh3
|
cadherin 3 |
| chr4_-_49845549 | 0.18 |
ENSMUST00000093859.11
ENSMUST00000076674.4 |
Grin3a
|
glutamate receptor ionotropic, NMDA3A |
| chr1_+_158190090 | 0.18 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chrX_+_161543384 | 0.18 |
ENSMUST00000033720.12
ENSMUST00000112327.8 |
Rbbp7
|
retinoblastoma binding protein 7, chromatin remodeling factor |
| chr4_-_56990306 | 0.18 |
ENSMUST00000053681.6
|
Frrs1l
|
ferric-chelate reductase 1 like |
| chr7_-_142522995 | 0.18 |
ENSMUST00000009392.6
|
Ascl2
|
achaete-scute family bHLH transcription factor 2 |
| chrX_+_72800675 | 0.18 |
ENSMUST00000002079.7
|
Plxnb3
|
plexin B3 |
| chrX_+_56008685 | 0.17 |
ENSMUST00000096431.10
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr15_-_79976016 | 0.17 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr14_+_54713703 | 0.17 |
ENSMUST00000164697.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr17_-_71158184 | 0.17 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr11_-_28533995 | 0.17 |
ENSMUST00000146385.9
|
Ccdc85a
|
coiled-coil domain containing 85A |
| chr7_+_28937859 | 0.17 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr2_+_128971620 | 0.17 |
ENSMUST00000035481.5
|
Chchd5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr11_+_115310885 | 0.17 |
ENSMUST00000103035.10
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.5 | 1.9 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.5 | 1.4 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.4 | 1.5 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.3 | 1.7 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.3 | 0.9 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.3 | 1.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.3 | 1.3 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.2 | 2.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 1.6 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 0.8 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.7 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 2.4 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 0.6 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 1.7 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.4 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.1 | 0.6 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 1.0 | GO:1901750 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 1.2 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.4 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 3.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.1 | 0.3 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.1 | 0.3 | GO:1903167 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) glycolate biosynthetic process(GO:0046295) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 0.2 | GO:0002660 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.5 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.9 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.7 | GO:1903797 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 0.4 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 1.1 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.1 | 0.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.7 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 0.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.1 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 1.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.3 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0060460 | subthalamic nucleus development(GO:0021763) left lung morphogenesis(GO:0060460) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 1.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.0 | 0.8 | GO:0008535 | cellular copper ion homeostasis(GO:0006878) respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.0 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.3 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 2.2 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.6 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.7 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.8 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.4 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:2000836 | positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.5 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.4 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 0.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.4 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.3 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.4 | GO:0000805 | X chromosome(GO:0000805) |
| 0.1 | 0.3 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 1.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0044279 | growing cell tip(GO:0035838) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.0 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.3 | 0.8 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.2 | 1.6 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.2 | 0.9 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.9 | GO:0031720 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.2 | 0.5 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 0.5 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 1.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 2.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.1 | 0.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 1.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.3 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 1.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 1.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 2.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0031402 | voltage-gated sodium channel activity(GO:0005248) sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 4.4 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 3.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.9 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 0.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |