Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
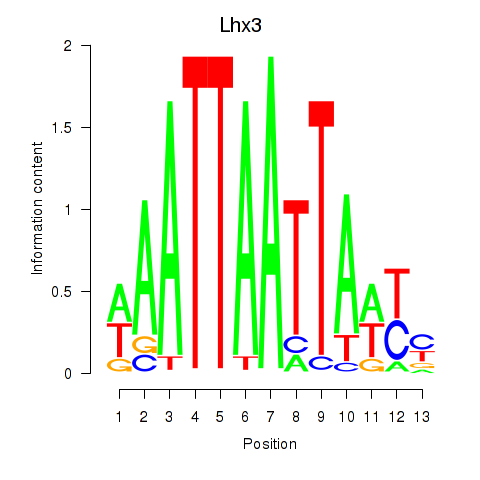
Results for Lhx3
Z-value: 0.52
Transcription factors associated with Lhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx3
|
ENSMUSG00000026934.16 | LIM homeobox protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx3 | mm39_v1_chr2_-_26098293_26098305 | -0.26 | 1.3e-01 | Click! |
Activity profile of Lhx3 motif
Sorted Z-values of Lhx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_20579322 | 5.36 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr3_+_59989282 | 4.07 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr3_-_67422821 | 2.59 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr18_-_38999755 | 2.17 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr1_+_21310803 | 1.70 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr1_+_21310821 | 1.65 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr7_+_107166925 | 1.61 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr2_+_22959223 | 1.52 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr9_-_110576192 | 1.45 |
ENSMUST00000199791.2
|
Pth1r
|
parathyroid hormone 1 receptor |
| chrM_+_9870 | 1.37 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chrM_+_9459 | 1.35 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr11_+_70410445 | 1.29 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chrM_+_14138 | 1.19 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr18_-_39000056 | 1.19 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr3_+_95226093 | 1.10 |
ENSMUST00000139866.2
|
Cers2
|
ceramide synthase 2 |
| chr2_+_22959452 | 1.06 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr9_+_21634779 | 0.98 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chrM_-_14061 | 0.90 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr5_+_96357974 | 0.84 |
ENSMUST00000036437.13
ENSMUST00000121477.2 |
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr15_+_65682066 | 0.76 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr3_+_151143524 | 0.75 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr17_-_35081129 | 0.75 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr17_-_35081456 | 0.75 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr17_-_36343573 | 0.74 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr16_+_11224481 | 0.73 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr3_+_122213420 | 0.73 |
ENSMUST00000029766.9
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr6_-_115569504 | 0.67 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr4_+_104770653 | 0.63 |
ENSMUST00000106803.9
ENSMUST00000106804.2 |
Fyb2
|
FYN binding protein 2 |
| chr5_+_96357337 | 0.62 |
ENSMUST00000117766.8
|
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr5_-_3697806 | 0.61 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr12_-_25147139 | 0.60 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr4_+_100336003 | 0.58 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr9_-_110576124 | 0.58 |
ENSMUST00000199862.5
ENSMUST00000198865.5 |
Pth1r
|
parathyroid hormone 1 receptor |
| chr11_-_49004584 | 0.57 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr17_+_79919267 | 0.55 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr6_+_34757346 | 0.55 |
ENSMUST00000115016.8
ENSMUST00000115017.8 |
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr9_-_15212745 | 0.55 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr2_+_124978518 | 0.52 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr16_-_64591509 | 0.52 |
ENSMUST00000076991.7
|
4930453N24Rik
|
RIKEN cDNA 4930453N24 gene |
| chr10_-_33500583 | 0.49 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr3_+_151143557 | 0.49 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr7_-_44753168 | 0.47 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr3_+_132335704 | 0.47 |
ENSMUST00000212594.2
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr12_-_84664001 | 0.46 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr14_-_63221950 | 0.45 |
ENSMUST00000100493.3
|
Defb48
|
defensin beta 48 |
| chr19_-_39801188 | 0.45 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr11_+_109434519 | 0.44 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr4_-_14621805 | 0.43 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr7_-_12829100 | 0.42 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr4_-_14621669 | 0.41 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr2_+_81883566 | 0.41 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr11_-_4045343 | 0.40 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr11_-_99979052 | 0.40 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr15_+_98350469 | 0.40 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr5_-_118382926 | 0.39 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr15_-_103473481 | 0.38 |
ENSMUST00000228060.2
ENSMUST00000228895.2 ENSMUST00000023134.5 |
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr2_-_32977182 | 0.36 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chrX_-_142716085 | 0.36 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr2_+_124978612 | 0.35 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr14_+_69585036 | 0.34 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr2_+_111607774 | 0.33 |
ENSMUST00000214708.2
ENSMUST00000215244.2 |
Olfr1302
|
olfactory receptor 1302 |
| chr9_+_120758282 | 0.32 |
ENSMUST00000130466.8
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr4_+_102446883 | 0.32 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr5_-_104225458 | 0.31 |
ENSMUST00000198485.5
ENSMUST00000164471.8 ENSMUST00000178967.2 |
Gm17660
|
predicted gene, 17660 |
| chr3_+_132335575 | 0.31 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr11_-_101066266 | 0.30 |
ENSMUST00000062759.4
|
Ccr10
|
chemokine (C-C motif) receptor 10 |
| chr13_+_93440572 | 0.30 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr5_+_143166759 | 0.30 |
ENSMUST00000031574.10
|
Spdye4b
|
speedy/RINGO cell cycle regulator family, member E4B |
| chr11_+_59197746 | 0.29 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr6_-_70313491 | 0.28 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr7_-_37472290 | 0.28 |
ENSMUST00000176205.8
|
Zfp536
|
zinc finger protein 536 |
| chr15_+_98468885 | 0.27 |
ENSMUST00000023728.8
|
Tex49
|
testis expressed 49 |
| chr1_+_88234454 | 0.27 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr3_+_135053762 | 0.26 |
ENSMUST00000159658.8
ENSMUST00000078568.12 |
Slc9b1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr2_-_32976378 | 0.26 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr4_-_14621497 | 0.25 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr1_+_177272215 | 0.25 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chrX_-_142716200 | 0.25 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr9_+_40092216 | 0.25 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr16_-_29363671 | 0.25 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr17_+_37977879 | 0.24 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr6_-_70116066 | 0.24 |
ENSMUST00000103379.3
ENSMUST00000197371.2 |
Igkv6-29
|
immunoglobulin kappa chain variable 6-29 |
| chr8_+_84262409 | 0.23 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr9_+_18320390 | 0.22 |
ENSMUST00000098973.3
|
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr10_+_36382810 | 0.22 |
ENSMUST00000167191.8
ENSMUST00000058738.11 |
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr4_+_145595364 | 0.22 |
ENSMUST00000123460.2
|
Zfp986
|
zinc finger protein 986 |
| chr2_-_164013033 | 0.21 |
ENSMUST00000045196.4
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chrX_+_65696608 | 0.21 |
ENSMUST00000036043.5
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr13_-_43634695 | 0.21 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr5_-_74863514 | 0.21 |
ENSMUST00000117388.8
|
Lnx1
|
ligand of numb-protein X 1 |
| chr7_-_44752508 | 0.21 |
ENSMUST00000209830.2
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr2_-_111779785 | 0.20 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr4_+_145241454 | 0.20 |
ENSMUST00000105741.2
|
Zfp990
|
zinc finger protein 990 |
| chr7_+_43077088 | 0.20 |
ENSMUST00000239023.2
|
Gm38999
|
predicted gene, 38999 |
| chr14_-_109151590 | 0.20 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr7_+_126575752 | 0.19 |
ENSMUST00000206346.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr10_-_8638215 | 0.19 |
ENSMUST00000212553.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr1_+_177272297 | 0.18 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr4_+_138606671 | 0.18 |
ENSMUST00000105804.2
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr3_+_41697046 | 0.18 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr6_-_128252540 | 0.17 |
ENSMUST00000130454.8
|
Tead4
|
TEA domain family member 4 |
| chr10_-_23226684 | 0.15 |
ENSMUST00000220299.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr3_-_64473251 | 0.15 |
ENSMUST00000176481.9
|
Vmn2r6
|
vomeronasal 2, receptor 6 |
| chr2_+_89757653 | 0.15 |
ENSMUST00000213720.3
ENSMUST00000102609.3 |
Olfr1258
|
olfactory receptor 1258 |
| chr4_+_147576874 | 0.15 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chr2_+_88644840 | 0.15 |
ENSMUST00000214703.2
|
Olfr1202
|
olfactory receptor 1202 |
| chr1_+_153541412 | 0.14 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr13_+_23343482 | 0.14 |
ENSMUST00000226845.2
ENSMUST00000228666.2 ENSMUST00000227388.2 |
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chr5_-_142515792 | 0.14 |
ENSMUST00000099400.3
|
Papolb
|
poly (A) polymerase beta (testis specific) |
| chr1_+_153541339 | 0.14 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr6_+_37847721 | 0.14 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr2_-_89855921 | 0.13 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr11_+_58485940 | 0.13 |
ENSMUST00000214990.2
ENSMUST00000216965.2 |
Olfr324
|
olfactory receptor 324 |
| chr13_+_93441307 | 0.13 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr19_+_26600820 | 0.13 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_+_53574579 | 0.12 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr10_+_23672842 | 0.12 |
ENSMUST00000119597.8
ENSMUST00000179321.8 ENSMUST00000133289.2 |
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr2_-_89678487 | 0.12 |
ENSMUST00000214428.3
|
Olfr48
|
olfactory receptor 48 |
| chr4_-_3938352 | 0.12 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr17_-_78991691 | 0.12 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr4_+_146033882 | 0.11 |
ENSMUST00000105730.2
ENSMUST00000091878.6 |
Zfp987
|
zinc finger protein 987 |
| chr18_+_37610858 | 0.11 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr4_+_145237329 | 0.11 |
ENSMUST00000105742.8
ENSMUST00000136309.8 |
Zfp990
|
zinc finger protein 990 |
| chr11_+_58311921 | 0.11 |
ENSMUST00000013797.3
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chr18_+_57601541 | 0.11 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr1_-_158183894 | 0.11 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr3_-_20421288 | 0.11 |
ENSMUST00000163776.3
ENSMUST00000068316.8 |
Agtr1b
|
angiotensin II receptor, type 1b |
| chr8_-_22396428 | 0.11 |
ENSMUST00000051965.5
|
Defb11
|
defensin beta 11 |
| chr5_-_70999547 | 0.10 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr11_-_83959999 | 0.10 |
ENSMUST00000138208.2
|
Dusp14
|
dual specificity phosphatase 14 |
| chr4_+_147056433 | 0.10 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989 |
| chr17_+_69746321 | 0.10 |
ENSMUST00000169935.2
|
Akain1
|
A kinase (PRKA) anchor inhibitor 1 |
| chr5_-_62923463 | 0.10 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_+_118818775 | 0.10 |
ENSMUST00000058651.5
|
Lao1
|
L-amino acid oxidase 1 |
| chr4_+_88768324 | 0.10 |
ENSMUST00000094972.2
|
Ifna1
|
interferon alpha 1 |
| chr7_-_103191732 | 0.09 |
ENSMUST00000215663.2
|
Olfr612
|
olfactory receptor 612 |
| chr5_-_51711204 | 0.09 |
ENSMUST00000196968.5
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr6_+_96092230 | 0.09 |
ENSMUST00000075080.6
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr4_-_146993984 | 0.09 |
ENSMUST00000238583.2
ENSMUST00000049821.4 |
Gm21411
|
predicted gene, 21411 |
| chr4_+_145397238 | 0.09 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr10_-_63926044 | 0.08 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr16_-_19341016 | 0.08 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr17_+_37769807 | 0.08 |
ENSMUST00000214668.2
ENSMUST00000217602.2 ENSMUST00000214938.2 |
Olfr109
|
olfactory receptor 109 |
| chr10_-_130265572 | 0.07 |
ENSMUST00000171811.4
|
Vmn2r85
|
vomeronasal 2, receptor 85 |
| chr5_-_51711237 | 0.06 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr17_-_68257900 | 0.06 |
ENSMUST00000164647.8
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr19_-_12313274 | 0.06 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr13_-_97170849 | 0.05 |
ENSMUST00000091377.5
|
1700029F12Rik
|
RIKEN cDNA 1700029F12 gene |
| chr8_+_22329942 | 0.05 |
ENSMUST00000006745.4
|
Defb2
|
defensin beta 2 |
| chr4_-_147787010 | 0.05 |
ENSMUST00000117638.2
|
Zfp534
|
zinc finger protein 534 |
| chr2_-_73410632 | 0.04 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr11_+_73489420 | 0.04 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr14_-_50586329 | 0.04 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr14_-_96756503 | 0.04 |
ENSMUST00000022666.9
|
Klhl1
|
kelch-like 1 |
| chr2_+_89708781 | 0.04 |
ENSMUST00000111519.3
|
Olfr1257
|
olfactory receptor 1257 |
| chr7_-_11414074 | 0.03 |
ENSMUST00000227010.2
ENSMUST00000209638.3 |
Vmn1r72
|
vomeronasal 1 receptor 72 |
| chr6_-_66537080 | 0.03 |
ENSMUST00000079584.3
ENSMUST00000227014.2 |
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr4_+_147390131 | 0.03 |
ENSMUST00000148762.4
|
Zfp988
|
zinc finger protein 988 |
| chr16_+_58967409 | 0.01 |
ENSMUST00000216957.3
|
Olfr195
|
olfactory receptor 195 |
| chr17_-_37615204 | 0.01 |
ENSMUST00000214376.2
|
Olfr101
|
olfactory receptor 101 |
| chr2_-_89779008 | 0.01 |
ENSMUST00000214846.2
|
Olfr1259
|
olfactory receptor 1259 |
| chr5_+_67126304 | 0.01 |
ENSMUST00000132991.5
|
Limch1
|
LIM and calponin homology domains 1 |
| chr14_-_48902555 | 0.01 |
ENSMUST00000118578.9
|
Otx2
|
orthodenticle homeobox 2 |
| chr7_+_24335969 | 0.01 |
ENSMUST00000080718.6
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr9_+_111014254 | 0.01 |
ENSMUST00000198986.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_+_181632922 | 0.00 |
ENSMUST00000071760.8
ENSMUST00000236373.2 ENSMUST00000184507.3 |
Gm14496
|
predicted gene 14496 |
| chr6_-_3399451 | 0.00 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
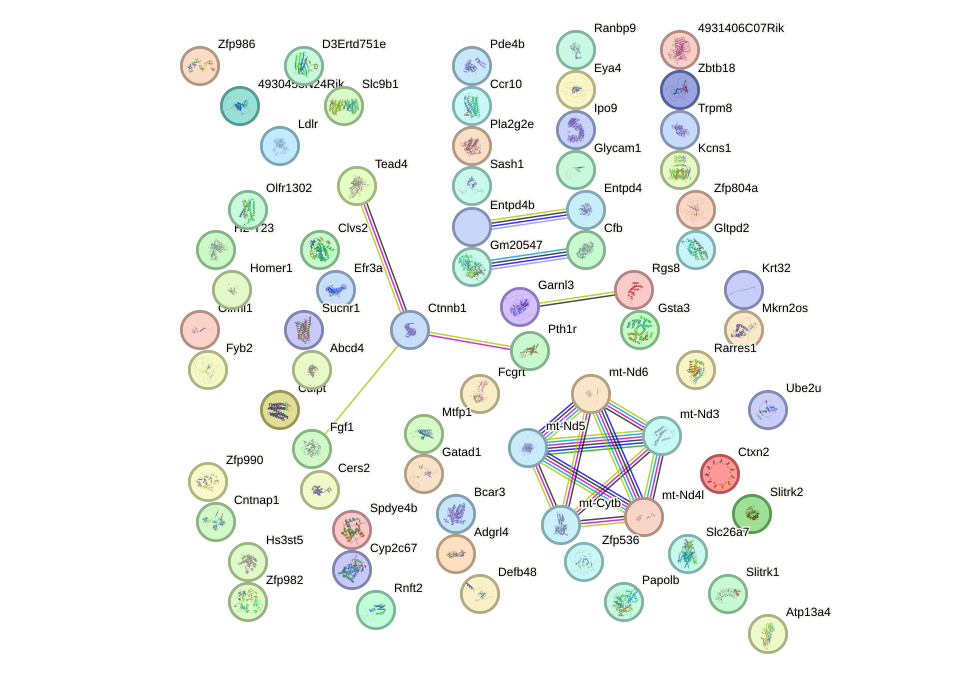
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.6 | 5.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 3.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 1.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 0.7 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.2 | 1.0 | GO:1903978 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) regulation of microglial cell activation(GO:1903978) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 1.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 2.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 0.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 1.2 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 1.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:1904499 | canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 2.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 1.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.6 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.2 | GO:1903210 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 1.5 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 1.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 1.5 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.2 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.3 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 2.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.2 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.0 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 2.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 2.0 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 1.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 3.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 3.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 2.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 3.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.7 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.4 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.3 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 3.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |