Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
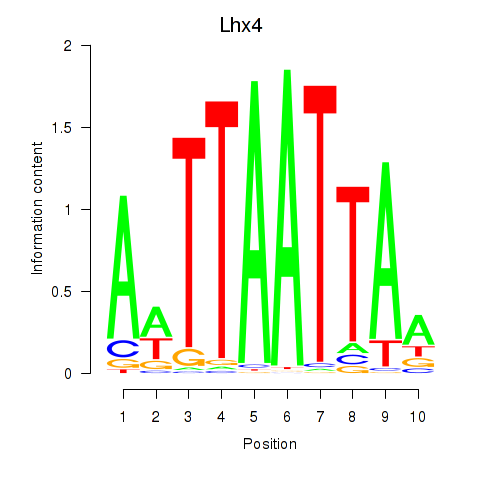
Results for Lhx4
Z-value: 0.51
Transcription factors associated with Lhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx4
|
ENSMUSG00000026468.15 | LIM homeobox protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx4 | mm39_v1_chr1_-_155617773_155617790 | -0.05 | 7.5e-01 | Click! |
Activity profile of Lhx4 motif
Sorted Z-values of Lhx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_80237691 | 5.98 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr11_+_32236945 | 4.13 |
ENSMUST00000101387.4
|
Hbq1b
|
hemoglobin, theta 1B |
| chr4_+_114914880 | 2.83 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr15_+_82225380 | 2.67 |
ENSMUST00000050349.3
|
Pheta2
|
PH domain containing endocytic trafficking adaptor 2 |
| chrX_+_149330371 | 2.39 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr10_+_75402090 | 2.00 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr6_-_136918885 | 1.93 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chrX_-_73290140 | 1.59 |
ENSMUST00000101454.9
ENSMUST00000033699.13 |
Flna
|
filamin, alpha |
| chr6_-_41423004 | 1.59 |
ENSMUST00000095999.7
|
Gm10334
|
predicted gene 10334 |
| chr6_-_115014777 | 1.58 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr9_+_96140781 | 1.55 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr14_+_26722319 | 1.54 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chrX_-_138772383 | 1.53 |
ENSMUST00000033811.14
ENSMUST00000087401.12 |
Morc4
|
microrchidia 4 |
| chr19_-_46033353 | 1.49 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr6_-_41354538 | 1.43 |
ENSMUST00000096003.7
|
Prss3
|
protease, serine 3 |
| chrX_-_73289970 | 1.42 |
ENSMUST00000130007.8
|
Flna
|
filamin, alpha |
| chr9_+_118892497 | 1.33 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chr2_+_163500290 | 1.26 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr17_+_34811217 | 1.23 |
ENSMUST00000038149.13
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr16_-_21980200 | 1.19 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr10_+_79832313 | 1.15 |
ENSMUST00000132517.8
|
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr3_-_14843512 | 1.06 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr7_+_30193047 | 0.99 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr5_+_35156389 | 0.94 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr17_+_46961250 | 0.94 |
ENSMUST00000043464.14
|
Cul7
|
cullin 7 |
| chr3_-_86827640 | 0.93 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr9_+_64188857 | 0.92 |
ENSMUST00000215031.2
ENSMUST00000213165.2 ENSMUST00000213289.2 ENSMUST00000216594.2 ENSMUST00000034964.7 |
Tipin
|
timeless interacting protein |
| chr5_-_122959321 | 0.89 |
ENSMUST00000197074.5
ENSMUST00000199406.5 ENSMUST00000196640.5 ENSMUST00000197719.5 ENSMUST00000200645.5 |
Anapc5
|
anaphase-promoting complex subunit 5 |
| chr3_-_86827664 | 0.87 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr7_-_45480200 | 0.86 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr1_+_34511793 | 0.84 |
ENSMUST00000188972.3
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr19_+_5524701 | 0.84 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr4_+_34893772 | 0.83 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr3_+_89622323 | 0.82 |
ENSMUST00000098924.9
|
Adar
|
adenosine deaminase, RNA-specific |
| chr9_+_96140750 | 0.81 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr10_-_35587888 | 0.80 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chrX_-_101200670 | 0.80 |
ENSMUST00000056904.3
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr13_+_20274708 | 0.80 |
ENSMUST00000072519.7
|
Elmo1
|
engulfment and cell motility 1 |
| chr2_+_36120438 | 0.78 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr6_+_123239076 | 0.75 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr15_-_103123711 | 0.73 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr7_-_108774367 | 0.72 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr1_+_63216281 | 0.69 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr6_+_72074718 | 0.68 |
ENSMUST00000187007.3
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr17_-_56343531 | 0.68 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr17_-_56343625 | 0.68 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr6_+_72074545 | 0.66 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr9_+_21437440 | 0.65 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chr3_-_88317601 | 0.64 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr5_+_35156454 | 0.63 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr1_-_36312482 | 0.62 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr1_+_63215976 | 0.61 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr17_-_24292453 | 0.61 |
ENSMUST00000017090.6
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chrX_+_152506577 | 0.61 |
ENSMUST00000140575.8
ENSMUST00000208373.2 ENSMUST00000185492.7 ENSMUST00000149514.8 |
Nbdy
|
negative regulator of P-body association |
| chr15_-_98507913 | 0.60 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr19_-_39875192 | 0.57 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr6_-_30936013 | 0.56 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr1_+_85503397 | 0.54 |
ENSMUST00000178024.2
|
G530012D18Rik
|
RIKEN cDNA G530012D1 gene |
| chr7_-_5017642 | 0.54 |
ENSMUST00000207412.2
ENSMUST00000077385.15 ENSMUST00000165320.3 |
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chr10_-_37014859 | 0.50 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr7_-_115459082 | 0.50 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr19_+_46385321 | 0.50 |
ENSMUST00000039922.13
ENSMUST00000111867.9 ENSMUST00000120778.8 |
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr3_-_72875187 | 0.49 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr7_-_37472979 | 0.47 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr10_-_75946790 | 0.47 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr3_+_68776884 | 0.47 |
ENSMUST00000054551.3
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr18_+_52958382 | 0.46 |
ENSMUST00000238707.2
|
Gm50457
|
predicted gene, 50457 |
| chr2_-_174188505 | 0.46 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr7_+_45271229 | 0.45 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chrX_+_132751729 | 0.44 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr14_+_57762191 | 0.44 |
ENSMUST00000089494.6
|
Il17d
|
interleukin 17D |
| chr2_-_88157559 | 0.43 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chrX_+_9751861 | 0.42 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr6_+_136509922 | 0.42 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr2_+_109522781 | 0.40 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr7_-_15781838 | 0.37 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chrX_-_8118541 | 0.37 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr11_+_6510167 | 0.37 |
ENSMUST00000109722.9
|
Ccm2
|
cerebral cavernous malformation 2 |
| chr2_+_85838122 | 0.33 |
ENSMUST00000062166.2
|
Olfr1032
|
olfactory receptor 1032 |
| chr7_+_89814713 | 0.33 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chrX_+_168468186 | 0.33 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr2_+_87696836 | 0.33 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr2_-_34716199 | 0.32 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr16_-_90607251 | 0.32 |
ENSMUST00000140920.2
|
Urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr2_-_111843053 | 0.31 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr9_-_49710190 | 0.31 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr6_-_141719536 | 0.30 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr10_-_23112973 | 0.30 |
ENSMUST00000218049.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr9_-_49710058 | 0.30 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr10_+_102376109 | 0.30 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr15_+_34453432 | 0.29 |
ENSMUST00000060894.9
|
Erich5
|
glutamate rich 5 |
| chr10_+_101994841 | 0.29 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr8_+_66964401 | 0.29 |
ENSMUST00000002025.5
ENSMUST00000183187.2 |
Tktl2
|
transketolase-like 2 |
| chr5_-_53864595 | 0.29 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr7_+_126575510 | 0.29 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr12_-_118930130 | 0.28 |
ENSMUST00000035515.5
|
Abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr4_+_146586445 | 0.28 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr4_+_134658209 | 0.28 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr10_+_74872898 | 0.28 |
ENSMUST00000147802.9
ENSMUST00000020391.13 ENSMUST00000234625.2 |
Rab36
|
RAB36, member RAS oncogene family |
| chr5_+_107656810 | 0.28 |
ENSMUST00000160160.6
|
Gm42669
|
predicted gene 42669 |
| chr4_+_148985584 | 0.27 |
ENSMUST00000147270.2
|
Casz1
|
castor zinc finger 1 |
| chr2_+_86122799 | 0.27 |
ENSMUST00000217166.2
|
Olfr1052
|
olfactory receptor 1052 |
| chr12_+_38833501 | 0.27 |
ENSMUST00000159334.8
|
Etv1
|
ets variant 1 |
| chr14_+_8283087 | 0.27 |
ENSMUST00000206298.3
ENSMUST00000216079.2 |
Olfr720
|
olfactory receptor 720 |
| chr4_+_145311759 | 0.27 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268 |
| chr12_+_59060162 | 0.27 |
ENSMUST00000021379.8
|
Gemin2
|
gem nuclear organelle associated protein 2 |
| chr12_+_38833454 | 0.26 |
ENSMUST00000161980.8
ENSMUST00000160701.8 |
Etv1
|
ets variant 1 |
| chr11_+_76795292 | 0.26 |
ENSMUST00000142166.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr16_+_58967409 | 0.26 |
ENSMUST00000216957.3
|
Olfr195
|
olfactory receptor 195 |
| chr5_-_86893645 | 0.26 |
ENSMUST00000161306.2
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr2_-_86944911 | 0.26 |
ENSMUST00000216088.3
|
Olfr259
|
olfactory receptor 259 |
| chr12_-_111947487 | 0.26 |
ENSMUST00000190536.2
|
Rd3l
|
retinal degeneration 3-like |
| chr14_-_110992533 | 0.26 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr8_+_13919641 | 0.25 |
ENSMUST00000051870.8
ENSMUST00000128557.3 |
Champ1
|
chromosome alignment maintaining phosphoprotein 1 |
| chr10_-_40178182 | 0.25 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr2_-_34716083 | 0.24 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr12_-_111947536 | 0.24 |
ENSMUST00000185354.2
|
Rd3l
|
retinal degeneration 3-like |
| chr7_+_99808452 | 0.24 |
ENSMUST00000032967.4
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr8_-_79539838 | 0.24 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr8_-_62576140 | 0.24 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr2_-_111965322 | 0.24 |
ENSMUST00000213696.2
|
Olfr1316
|
olfactory receptor 1316 |
| chr3_+_138058139 | 0.24 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr2_-_89779008 | 0.23 |
ENSMUST00000214846.2
|
Olfr1259
|
olfactory receptor 1259 |
| chr19_-_24178000 | 0.23 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr11_+_76795346 | 0.23 |
ENSMUST00000072633.4
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chrX_+_56257374 | 0.23 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr11_+_31823096 | 0.23 |
ENSMUST00000155278.2
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr6_-_69282389 | 0.22 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr2_-_73284262 | 0.22 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr2_-_87570322 | 0.22 |
ENSMUST00000214573.2
|
Olfr1138
|
olfactory receptor 1138 |
| chr14_-_50479161 | 0.22 |
ENSMUST00000214388.2
|
Olfr731
|
olfactory receptor 731 |
| chr4_-_45532470 | 0.21 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr4_-_58499398 | 0.20 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr4_+_147216495 | 0.20 |
ENSMUST00000084149.10
|
Zfp991
|
zinc finger protein 991 |
| chr16_-_26786995 | 0.20 |
ENSMUST00000231969.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr2_-_86257093 | 0.20 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr19_-_55229668 | 0.20 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr13_-_52685305 | 0.20 |
ENSMUST00000057442.8
|
Diras2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr7_+_99808526 | 0.20 |
ENSMUST00000207825.2
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr17_+_29493113 | 0.20 |
ENSMUST00000234326.2
ENSMUST00000235117.2 |
BC004004
|
cDNA sequence BC004004 |
| chr10_-_130230201 | 0.20 |
ENSMUST00000094502.6
ENSMUST00000233146.2 |
Vmn2r84
|
vomeronasal 2, receptor 84 |
| chr6_-_136899167 | 0.20 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr2_+_85648823 | 0.20 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr4_+_11558905 | 0.19 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr9_-_103569984 | 0.19 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr14_+_47710574 | 0.19 |
ENSMUST00000228740.2
|
Fbxo34
|
F-box protein 34 |
| chr17_+_17622934 | 0.19 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr14_+_8282925 | 0.19 |
ENSMUST00000217642.2
|
Olfr720
|
olfactory receptor 720 |
| chr2_-_79287095 | 0.19 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr14_-_104760051 | 0.18 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr11_-_73348284 | 0.18 |
ENSMUST00000121209.3
ENSMUST00000127789.3 |
Olfr380
|
olfactory receptor 380 |
| chr9_+_108216233 | 0.17 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr8_-_126296495 | 0.17 |
ENSMUST00000212168.2
ENSMUST00000212987.2 |
Sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr10_+_101994719 | 0.17 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr17_+_29493049 | 0.17 |
ENSMUST00000149405.4
|
BC004004
|
cDNA sequence BC004004 |
| chr2_-_88562710 | 0.17 |
ENSMUST00000213118.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr19_-_12313274 | 0.17 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr11_-_99433984 | 0.17 |
ENSMUST00000107443.8
ENSMUST00000074253.4 |
Krt40
|
keratin 40 |
| chr11_+_62737887 | 0.17 |
ENSMUST00000036085.11
|
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr4_-_147894245 | 0.17 |
ENSMUST00000105734.10
ENSMUST00000176201.2 |
Zfp984
Gm20707
|
zinc finger protein 984 predicted gene 20707 |
| chr3_-_96359622 | 0.17 |
ENSMUST00000093126.11
ENSMUST00000098841.4 |
BC107364
|
cDNA sequence BC107364 |
| chr11_+_58062467 | 0.17 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr1_-_4430481 | 0.16 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr14_+_53994813 | 0.16 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr4_+_145397238 | 0.16 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr6_+_29859685 | 0.15 |
ENSMUST00000134438.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr7_+_84562283 | 0.15 |
ENSMUST00000216367.2
ENSMUST00000214501.3 |
Olfr290
|
olfactory receptor 290 |
| chr19_-_13828056 | 0.15 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr14_+_27598021 | 0.15 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr15_+_16778187 | 0.15 |
ENSMUST00000026432.8
|
Cdh9
|
cadherin 9 |
| chr11_+_62737936 | 0.15 |
ENSMUST00000150989.8
ENSMUST00000176577.2 |
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr6_+_29859660 | 0.15 |
ENSMUST00000128927.9
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr19_-_45224251 | 0.15 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chrX_-_74621828 | 0.15 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr5_+_13448833 | 0.15 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_-_165462020 | 0.14 |
ENSMUST00000194437.6
ENSMUST00000068705.13 ENSMUST00000111435.9 ENSMUST00000193023.2 |
Mpzl1
|
myelin protein zero-like 1 |
| chr2_+_85545763 | 0.14 |
ENSMUST00000216443.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr7_+_107679062 | 0.14 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
| chr17_+_29493157 | 0.14 |
ENSMUST00000234234.2
|
BC004004
|
cDNA sequence BC004004 |
| chr18_+_57601541 | 0.14 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr15_-_56557920 | 0.14 |
ENSMUST00000050544.8
|
Has2
|
hyaluronan synthase 2 |
| chr6_-_145811028 | 0.14 |
ENSMUST00000111703.2
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr2_+_111084861 | 0.14 |
ENSMUST00000218065.2
|
Olfr1276
|
olfactory receptor 1276 |
| chr9_-_19275301 | 0.14 |
ENSMUST00000214810.2
|
Olfr846
|
olfactory receptor 846 |
| chr16_-_92196954 | 0.14 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr4_+_146695418 | 0.13 |
ENSMUST00000130825.8
|
Zfp993
|
zinc finger protein 993 |
| chr17_+_70829050 | 0.13 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr10_+_87041814 | 0.13 |
ENSMUST00000189775.2
|
1700113H08Rik
|
RIKEN cDNA 1700113H08 gene |
| chr2_-_87504008 | 0.13 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr8_+_121854566 | 0.13 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr19_-_13827773 | 0.13 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr10_+_115979787 | 0.12 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr10_+_129601351 | 0.12 |
ENSMUST00000203236.3
|
Olfr808
|
olfactory receptor 808 |
| chr2_-_7400690 | 0.12 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr6_+_96092230 | 0.12 |
ENSMUST00000075080.6
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr4_+_147637714 | 0.12 |
ENSMUST00000139784.8
ENSMUST00000143885.8 ENSMUST00000081742.7 |
Zfp985
|
zinc finger protein 985 |
| chr5_+_13448647 | 0.12 |
ENSMUST00000125629.8
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_14914484 | 0.12 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2 |
| chr4_+_147390131 | 0.12 |
ENSMUST00000148762.4
|
Zfp988
|
zinc finger protein 988 |
| chr4_+_145311722 | 0.12 |
ENSMUST00000105739.8
|
Zfp268
|
zinc finger protein 268 |
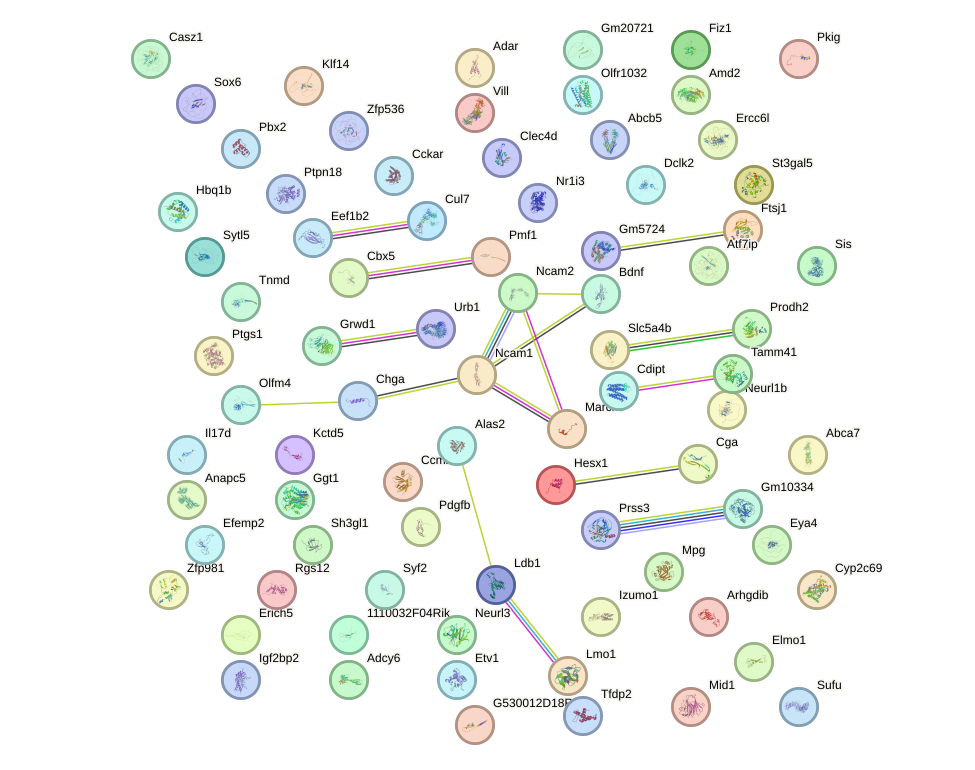
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.9 | 2.8 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.5 | 1.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.4 | 1.2 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.4 | 1.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 1.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 1.6 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.3 | 0.8 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.2 | 2.0 | GO:1901750 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 1.9 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 0.6 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.2 | 0.9 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 2.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 0.6 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 0.4 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 1.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.4 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 1.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.7 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.3 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 6.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.6 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 2.0 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.2 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.8 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.3 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.1 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.6 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.2 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 0.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.5 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.3 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 1.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 1.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 2.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 | 0.9 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.0 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.5 | 3.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 6.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 1.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.9 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 1.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.5 | 1.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.5 | 3.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.4 | 1.3 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.4 | 1.2 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 0.8 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.2 | 1.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 2.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 1.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0071820 | N-box binding(GO:0071820) |
| 0.0 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 3.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 1.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 1.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 5.7 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 3.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |