Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
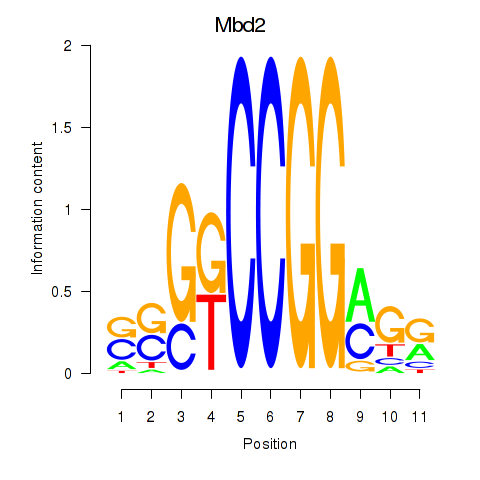
Results for Mbd2
Z-value: 0.95
Transcription factors associated with Mbd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mbd2
|
ENSMUSG00000024513.17 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mbd2 | mm39_v1_chr18_+_70701260_70701469 | -0.82 | 8.9e-10 | Click! |
Activity profile of Mbd2 motif
Sorted Z-values of Mbd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_65511777 | 3.90 |
ENSMUST00000055576.12
ENSMUST00000098391.11 |
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr12_+_87194476 | 2.80 |
ENSMUST00000063117.10
ENSMUST00000220574.2 |
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr12_-_102671154 | 2.70 |
ENSMUST00000178697.2
ENSMUST00000046518.12 |
Itpk1
|
inositol 1,3,4-triphosphate 5/6 kinase |
| chr5_-_53370761 | 2.59 |
ENSMUST00000031090.8
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr2_+_27567213 | 2.40 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr15_-_31367668 | 2.39 |
ENSMUST00000110410.10
ENSMUST00000076942.5 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr15_-_31367872 | 2.36 |
ENSMUST00000123325.9
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr8_-_71834543 | 2.24 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr7_+_86895851 | 2.18 |
ENSMUST00000032781.14
|
Nox4
|
NADPH oxidase 4 |
| chr7_+_65511482 | 2.12 |
ENSMUST00000176199.8
|
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr16_-_20534851 | 2.10 |
ENSMUST00000120099.8
ENSMUST00000232309.2 ENSMUST00000007207.15 |
Clcn2
|
chloride channel, voltage-sensitive 2 |
| chr16_-_10360893 | 2.08 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr3_+_89680867 | 2.00 |
ENSMUST00000038356.13
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr12_+_87193922 | 2.00 |
ENSMUST00000222885.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr13_-_53135064 | 2.00 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr15_-_85918378 | 1.87 |
ENSMUST00000016172.10
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr5_+_30971915 | 1.83 |
ENSMUST00000031058.15
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr2_+_27567246 | 1.82 |
ENSMUST00000166775.8
|
Rxra
|
retinoid X receptor alpha |
| chr17_+_34250757 | 1.82 |
ENSMUST00000044858.16
ENSMUST00000174299.9 |
Rxrb
|
retinoid X receptor beta |
| chr15_-_76540916 | 1.81 |
ENSMUST00000229524.2
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr5_+_30972067 | 1.81 |
ENSMUST00000200692.4
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr13_-_55510595 | 1.77 |
ENSMUST00000021940.8
|
Lman2
|
lectin, mannose-binding 2 |
| chr7_-_126625739 | 1.73 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr17_+_34251041 | 1.72 |
ENSMUST00000173354.9
|
Rxrb
|
retinoid X receptor beta |
| chr19_+_6291698 | 1.67 |
ENSMUST00000045351.13
|
Atg2a
|
autophagy related 2A |
| chr12_-_87194658 | 1.67 |
ENSMUST00000037788.6
|
Pomt2
|
protein-O-mannosyltransferase 2 |
| chr2_-_6327884 | 1.64 |
ENSMUST00000238876.2
|
1700014B07Rik
|
RIKEN cDNA 1700014B07 gene |
| chr3_-_121608809 | 1.56 |
ENSMUST00000197383.5
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr2_-_160714473 | 1.56 |
ENSMUST00000103111.9
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr4_-_155430153 | 1.54 |
ENSMUST00000103178.11
|
Prkcz
|
protein kinase C, zeta |
| chr5_+_54155814 | 1.53 |
ENSMUST00000117661.9
|
Stim2
|
stromal interaction molecule 2 |
| chr7_-_126625657 | 1.51 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr12_-_108241597 | 1.50 |
ENSMUST00000222310.2
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr11_-_89193158 | 1.49 |
ENSMUST00000061728.5
|
Nog
|
noggin |
| chr3_-_121608859 | 1.47 |
ENSMUST00000029770.8
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr7_-_127307898 | 1.47 |
ENSMUST00000207019.2
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr7_+_86895996 | 1.47 |
ENSMUST00000068829.13
|
Nox4
|
NADPH oxidase 4 |
| chr11_-_107685383 | 1.46 |
ENSMUST00000021066.4
|
Cacng4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr13_+_49340961 | 1.46 |
ENSMUST00000049022.15
|
Ninj1
|
ninjurin 1 |
| chr8_+_112263255 | 1.44 |
ENSMUST00000171182.8
ENSMUST00000168428.8 |
Znrf1
|
zinc and ring finger 1 |
| chr10_-_31321793 | 1.41 |
ENSMUST00000213639.2
ENSMUST00000215515.2 ENSMUST00000214644.2 ENSMUST00000213528.2 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr1_+_74430575 | 1.39 |
ENSMUST00000027367.14
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr12_+_17316546 | 1.39 |
ENSMUST00000057288.7
ENSMUST00000239402.2 |
Pdia6
|
protein disulfide isomerase associated 6 |
| chr12_-_107969673 | 1.38 |
ENSMUST00000109887.8
ENSMUST00000109891.3 |
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr5_+_54155871 | 1.38 |
ENSMUST00000201469.4
|
Stim2
|
stromal interaction molecule 2 |
| chr11_-_106811507 | 1.38 |
ENSMUST00000103067.10
|
Smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr7_+_29931309 | 1.37 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr13_+_49340995 | 1.37 |
ENSMUST00000120733.8
|
Ninj1
|
ninjurin 1 |
| chr2_+_78699360 | 1.37 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr15_+_87509413 | 1.32 |
ENSMUST00000068088.8
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr2_-_27365612 | 1.31 |
ENSMUST00000147736.2
|
Brd3
|
bromodomain containing 3 |
| chr14_+_122712809 | 1.30 |
ENSMUST00000075888.6
|
Zic2
|
zinc finger protein of the cerebellum 2 |
| chr7_-_127308059 | 1.29 |
ENSMUST00000061468.9
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr2_+_155849965 | 1.27 |
ENSMUST00000006035.13
|
Ergic3
|
ERGIC and golgi 3 |
| chr10_-_53951796 | 1.27 |
ENSMUST00000105470.9
|
Man1a
|
mannosidase 1, alpha |
| chr5_+_37025926 | 1.26 |
ENSMUST00000201156.2
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr2_+_155850002 | 1.25 |
ENSMUST00000088650.11
|
Ergic3
|
ERGIC and golgi 3 |
| chr7_-_74204222 | 1.25 |
ENSMUST00000134539.2
ENSMUST00000026897.14 ENSMUST00000098371.9 |
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr18_-_25301729 | 1.24 |
ENSMUST00000148255.8
|
Tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr9_-_110571645 | 1.23 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr4_-_3938352 | 1.22 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr8_+_112263632 | 1.22 |
ENSMUST00000173506.8
|
Znrf1
|
zinc and ring finger 1 |
| chr9_+_80072274 | 1.22 |
ENSMUST00000035889.15
ENSMUST00000113268.8 |
Myo6
|
myosin VI |
| chr8_+_112263465 | 1.21 |
ENSMUST00000095176.12
|
Znrf1
|
zinc and ring finger 1 |
| chr14_+_45457168 | 1.21 |
ENSMUST00000227086.2
ENSMUST00000147957.2 |
Gpr137c
|
G protein-coupled receptor 137C |
| chr11_-_86884507 | 1.21 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr14_-_20844074 | 1.19 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr18_+_84106188 | 1.17 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr2_+_155617251 | 1.16 |
ENSMUST00000029141.6
|
Mmp24
|
matrix metallopeptidase 24 |
| chr1_+_106099482 | 1.15 |
ENSMUST00000061047.7
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr11_+_87507977 | 1.15 |
ENSMUST00000093956.4
|
Hsf5
|
heat shock transcription factor family member 5 |
| chr9_+_43655230 | 1.12 |
ENSMUST00000034510.9
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr18_-_16942289 | 1.11 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr5_+_37025810 | 1.11 |
ENSMUST00000031003.11
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr4_-_120604445 | 1.09 |
ENSMUST00000030376.8
|
Kcnq4
|
potassium voltage-gated channel, subfamily Q, member 4 |
| chr10_-_31485180 | 1.08 |
ENSMUST00000081989.8
|
Rnf217
|
ring finger protein 217 |
| chr2_+_32536594 | 1.07 |
ENSMUST00000113272.8
ENSMUST00000009705.14 ENSMUST00000167841.8 |
Eng
|
endoglin |
| chr1_+_91729175 | 1.05 |
ENSMUST00000007949.4
ENSMUST00000186075.2 |
Twist2
|
twist basic helix-loop-helix transcription factor 2 |
| chr19_+_4147391 | 1.05 |
ENSMUST00000174514.2
ENSMUST00000174149.8 |
Cdk2ap2
|
CDK2-associated protein 2 |
| chr16_-_4238280 | 1.03 |
ENSMUST00000120080.8
|
Adcy9
|
adenylate cyclase 9 |
| chr8_-_123980825 | 1.02 |
ENSMUST00000118279.2
|
Vps9d1
|
VPS9 domain containing 1 |
| chr11_+_108811626 | 1.01 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr15_+_74435587 | 1.00 |
ENSMUST00000185682.7
ENSMUST00000170845.8 ENSMUST00000187599.2 |
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr4_+_152423344 | 0.99 |
ENSMUST00000005175.5
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr10_+_127216459 | 0.98 |
ENSMUST00000166820.8
|
R3hdm2
|
R3H domain containing 2 |
| chr2_-_94236991 | 0.98 |
ENSMUST00000111237.9
ENSMUST00000094801.5 ENSMUST00000111238.8 |
Ttc17
|
tetratricopeptide repeat domain 17 |
| chr2_-_155849839 | 0.96 |
ENSMUST00000086145.10
ENSMUST00000144686.8 ENSMUST00000140657.8 |
6430550D23Rik
|
RIKEN cDNA 6430550D23 gene |
| chr10_+_127216668 | 0.95 |
ENSMUST00000111628.9
|
R3hdm2
|
R3H domain containing 2 |
| chr12_-_108241392 | 0.95 |
ENSMUST00000136175.3
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr18_-_25302064 | 0.94 |
ENSMUST00000115817.3
|
Tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr7_-_127307791 | 0.92 |
ENSMUST00000205977.2
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr12_-_101942172 | 0.92 |
ENSMUST00000221191.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr10_-_77738300 | 0.91 |
ENSMUST00000057608.5
|
Lrrc3
|
leucine rich repeat containing 3 |
| chrX_+_160500623 | 0.91 |
ENSMUST00000061514.8
|
Rai2
|
retinoic acid induced 2 |
| chrX_+_150127171 | 0.90 |
ENSMUST00000073364.6
|
Fam120c
|
family with sequence similarity 120, member C |
| chr7_-_46782448 | 0.90 |
ENSMUST00000033142.13
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr14_-_70757601 | 0.89 |
ENSMUST00000022693.9
|
Bmp1
|
bone morphogenetic protein 1 |
| chr1_+_74640706 | 0.89 |
ENSMUST00000087186.11
|
Stk36
|
serine/threonine kinase 36 |
| chr8_-_123980908 | 0.88 |
ENSMUST00000122363.8
|
Vps9d1
|
VPS9 domain containing 1 |
| chr11_+_97554192 | 0.88 |
ENSMUST00000044730.12
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 6 |
| chr7_+_141503719 | 0.87 |
ENSMUST00000105989.9
ENSMUST00000075528.12 ENSMUST00000174499.8 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr17_+_83522700 | 0.87 |
ENSMUST00000170794.8
|
Pkdcc
|
protein kinase domain containing, cytoplasmic |
| chr1_+_91226058 | 0.86 |
ENSMUST00000027532.13
|
Scly
|
selenocysteine lyase |
| chr8_-_125161061 | 0.85 |
ENSMUST00000140012.8
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr14_-_39194782 | 0.85 |
ENSMUST00000168810.9
ENSMUST00000173780.2 ENSMUST00000166968.9 |
Nrg3
|
neuregulin 3 |
| chr10_-_81266800 | 0.84 |
ENSMUST00000117966.2
|
Nfic
|
nuclear factor I/C |
| chr2_+_157756535 | 0.84 |
ENSMUST00000109523.2
|
Vstm2l
|
V-set and transmembrane domain containing 2-like |
| chr6_-_122463316 | 0.84 |
ENSMUST00000205114.2
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr9_+_80072361 | 0.84 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr18_+_77861656 | 0.84 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr18_+_69479211 | 0.83 |
ENSMUST00000201235.4
|
Tcf4
|
transcription factor 4 |
| chr12_-_99359265 | 0.83 |
ENSMUST00000177451.8
|
Foxn3
|
forkhead box N3 |
| chr2_-_160714749 | 0.83 |
ENSMUST00000176141.8
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr4_-_43669141 | 0.82 |
ENSMUST00000056474.7
|
Fam221b
|
family with sequence similarity 221, member B |
| chr7_-_139162706 | 0.81 |
ENSMUST00000106095.3
|
Nkx6-2
|
NK6 homeobox 2 |
| chr1_+_74640590 | 0.80 |
ENSMUST00000087183.11
ENSMUST00000148456.8 ENSMUST00000113694.8 |
Stk36
|
serine/threonine kinase 36 |
| chr4_+_109835224 | 0.80 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr4_+_132766793 | 0.80 |
ENSMUST00000105914.2
|
Ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr11_+_116809669 | 0.80 |
ENSMUST00000103027.10
|
Mgat5b
|
mannoside acetylglucosaminyltransferase 5, isoenzyme B |
| chr14_-_29443792 | 0.79 |
ENSMUST00000022567.9
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr11_-_5394838 | 0.78 |
ENSMUST00000109867.8
ENSMUST00000143746.3 |
Znrf3
|
zinc and ring finger 3 |
| chr14_+_34542053 | 0.78 |
ENSMUST00000043349.7
|
Grid1
|
glutamate receptor, ionotropic, delta 1 |
| chr15_-_76541105 | 0.78 |
ENSMUST00000176274.2
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr19_+_8875459 | 0.78 |
ENSMUST00000096246.5
ENSMUST00000235274.2 |
Ganab
|
alpha glucosidase 2 alpha neutral subunit |
| chr7_+_4693603 | 0.77 |
ENSMUST00000120836.8
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr5_-_115332343 | 0.77 |
ENSMUST00000112113.8
|
Cabp1
|
calcium binding protein 1 |
| chr11_+_77928736 | 0.77 |
ENSMUST00000072289.12
ENSMUST00000100784.9 ENSMUST00000073660.7 |
Flot2
|
flotillin 2 |
| chr11_+_108811168 | 0.77 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr10_+_127000991 | 0.77 |
ENSMUST00000006914.11
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr7_+_141503411 | 0.76 |
ENSMUST00000078200.12
ENSMUST00000018971.15 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr5_-_24556602 | 0.76 |
ENSMUST00000036092.10
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr2_-_160714904 | 0.75 |
ENSMUST00000109460.8
ENSMUST00000127201.2 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr2_-_29142965 | 0.75 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr7_+_4693759 | 0.74 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr9_+_108569315 | 0.74 |
ENSMUST00000035220.12
|
Prkar2a
|
protein kinase, cAMP dependent regulatory, type II alpha |
| chr10_-_127502541 | 0.72 |
ENSMUST00000026469.9
|
Nab2
|
Ngfi-A binding protein 2 |
| chr2_-_166838018 | 0.72 |
ENSMUST00000049412.12
|
Stau1
|
staufen double-stranded RNA binding protein 1 |
| chr2_-_173118315 | 0.71 |
ENSMUST00000036248.13
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr17_+_35643853 | 0.70 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr6_+_124908439 | 0.70 |
ENSMUST00000032214.14
|
Mlf2
|
myeloid leukemia factor 2 |
| chr4_+_129878627 | 0.70 |
ENSMUST00000120204.8
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr15_+_89407954 | 0.70 |
ENSMUST00000230807.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr2_+_139520098 | 0.68 |
ENSMUST00000184404.8
ENSMUST00000099307.4 |
Ism1
|
isthmin 1, angiogenesis inhibitor |
| chr7_+_141503583 | 0.67 |
ENSMUST00000172652.8
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr2_-_166838182 | 0.67 |
ENSMUST00000109238.9
ENSMUST00000109235.8 ENSMUST00000109236.9 |
Stau1
|
staufen double-stranded RNA binding protein 1 |
| chr15_+_68800546 | 0.67 |
ENSMUST00000230847.2
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr1_-_165830160 | 0.67 |
ENSMUST00000111429.11
ENSMUST00000176800.2 ENSMUST00000177358.8 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr2_+_157120946 | 0.66 |
ENSMUST00000116380.9
ENSMUST00000029171.6 |
Rpn2
|
ribophorin II |
| chr2_+_179813278 | 0.66 |
ENSMUST00000061437.5
|
Adrm1
|
adhesion regulating molecule 1 |
| chr19_-_10502468 | 0.65 |
ENSMUST00000025570.8
ENSMUST00000236455.2 |
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr8_+_93687561 | 0.65 |
ENSMUST00000072939.8
|
Slc6a2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| chr11_-_35871300 | 0.65 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr9_+_26645141 | 0.64 |
ENSMUST00000115269.9
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr17_+_35643818 | 0.64 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr19_+_11943265 | 0.63 |
ENSMUST00000025590.11
|
Osbp
|
oxysterol binding protein |
| chr11_-_106378622 | 0.63 |
ENSMUST00000001059.9
ENSMUST00000106799.2 ENSMUST00000106800.2 |
Ern1
|
endoplasmic reticulum (ER) to nucleus signalling 1 |
| chr12_-_107969853 | 0.63 |
ENSMUST00000066060.11
|
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr19_-_47452557 | 0.63 |
ENSMUST00000111800.4
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr15_+_80118219 | 0.63 |
ENSMUST00000023048.12
|
Mief1
|
mitochondrial elongation factor 1 |
| chr19_-_10502546 | 0.63 |
ENSMUST00000237827.2
|
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr7_-_64806164 | 0.62 |
ENSMUST00000148459.3
ENSMUST00000119118.8 |
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr6_+_6863769 | 0.62 |
ENSMUST00000031768.8
|
Dlx6
|
distal-less homeobox 6 |
| chr4_+_43669266 | 0.62 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr12_-_111679379 | 0.62 |
ENSMUST00000160825.2
ENSMUST00000162953.2 |
Bag5
|
BCL2-associated athanogene 5 |
| chrX_+_160500051 | 0.62 |
ENSMUST00000112338.2
|
Rai2
|
retinoic acid induced 2 |
| chr1_-_38168801 | 0.61 |
ENSMUST00000195383.2
|
Rev1
|
REV1, DNA directed polymerase |
| chr7_-_125681577 | 0.61 |
ENSMUST00000073935.7
|
Gsg1l
|
GSG1-like |
| chr3_-_108133914 | 0.61 |
ENSMUST00000141387.4
|
Sypl2
|
synaptophysin-like 2 |
| chr11_-_120515799 | 0.61 |
ENSMUST00000106183.3
ENSMUST00000080202.12 |
Sirt7
|
sirtuin 7 |
| chr7_-_136915602 | 0.61 |
ENSMUST00000210774.2
|
Ebf3
|
early B cell factor 3 |
| chr2_-_84717036 | 0.61 |
ENSMUST00000054514.6
ENSMUST00000151799.8 |
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr18_+_69478893 | 0.60 |
ENSMUST00000202354.4
|
Tcf4
|
transcription factor 4 |
| chr7_+_29003363 | 0.60 |
ENSMUST00000108231.8
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr1_+_40720731 | 0.60 |
ENSMUST00000192345.2
|
Slc9a2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| chr17_-_16046780 | 0.60 |
ENSMUST00000232638.2
ENSMUST00000170578.3 |
Rgmb
|
repulsive guidance molecule family member B |
| chr15_+_34837501 | 0.60 |
ENSMUST00000072868.5
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr9_+_26645024 | 0.60 |
ENSMUST00000160899.8
ENSMUST00000161431.3 ENSMUST00000159799.8 |
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr8_-_88199231 | 0.59 |
ENSMUST00000034076.16
|
Cbln1
|
cerebellin 1 precursor protein |
| chr4_+_129878890 | 0.59 |
ENSMUST00000106017.8
ENSMUST00000121049.8 |
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr1_+_74894069 | 0.59 |
ENSMUST00000160379.4
|
Cdk5r2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr15_-_53209513 | 0.59 |
ENSMUST00000077273.9
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr15_+_74388044 | 0.59 |
ENSMUST00000042035.16
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr7_-_37470184 | 0.58 |
ENSMUST00000176680.8
|
Zfp536
|
zinc finger protein 536 |
| chr7_+_29979512 | 0.58 |
ENSMUST00000108187.8
ENSMUST00000014072.6 |
1700020L13Rik
|
RIKEN cDNA 1700020L13 gene |
| chr11_-_61157986 | 0.58 |
ENSMUST00000066277.10
ENSMUST00000074127.14 ENSMUST00000108715.3 |
Aldh3a2
|
aldehyde dehydrogenase family 3, subfamily A2 |
| chr2_+_156455583 | 0.57 |
ENSMUST00000109567.10
ENSMUST00000169464.9 |
Dlgap4
|
DLG associated protein 4 |
| chr5_+_114706077 | 0.57 |
ENSMUST00000043650.8
|
Fam222a
|
family with sequence similarity 222, member A |
| chr6_+_124908341 | 0.57 |
ENSMUST00000203021.3
|
Mlf2
|
myeloid leukemia factor 2 |
| chr4_-_57300361 | 0.57 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr7_-_99629637 | 0.57 |
ENSMUST00000080817.6
|
Rnf169
|
ring finger protein 169 |
| chr6_-_72876269 | 0.57 |
ENSMUST00000204598.3
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr4_+_152423075 | 0.56 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr5_-_138981842 | 0.56 |
ENSMUST00000110897.8
|
Pdgfa
|
platelet derived growth factor, alpha |
| chr5_-_108697857 | 0.56 |
ENSMUST00000129040.2
ENSMUST00000046892.10 |
Cplx1
|
complexin 1 |
| chr12_-_5425682 | 0.56 |
ENSMUST00000020958.9
|
Klhl29
|
kelch-like 29 |
| chr4_-_149858694 | 0.55 |
ENSMUST00000105686.3
|
Slc25a33
|
solute carrier family 25, member 33 |
| chr7_-_126625617 | 0.54 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr13_-_120252229 | 0.54 |
ENSMUST00000223812.2
|
Zfp131
|
zinc finger protein 131 |
| chr10_-_17898838 | 0.54 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr15_+_76215488 | 0.54 |
ENSMUST00000172281.8
|
Gpaa1
|
GPI anchor attachment protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mbd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.0 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.9 | 3.6 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.8 | 4.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.8 | 3.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.7 | 2.0 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.6 | 1.9 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.5 | 4.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 1.5 | GO:0060300 | regulation of cytokine activity(GO:0060300) fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.4 | 1.7 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.4 | 2.9 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.4 | 1.1 | GO:0003032 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.3 | 2.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.3 | 1.3 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.3 | 1.3 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.3 | 3.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 1.8 | GO:0061181 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.3 | 0.9 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.3 | 0.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 1.7 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.2 | 1.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 1.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 1.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 0.9 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.2 | 1.5 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 1.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.2 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.2 | 0.6 | GO:1903173 | sesquiterpenoid metabolic process(GO:0006714) phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.2 | 2.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.2 | 0.8 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.2 | 0.6 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.2 | 0.5 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.2 | 0.7 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 0.5 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 1.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 0.3 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 1.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.2 | 0.6 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 3.0 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 0.6 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 0.4 | GO:0015881 | creatine transport(GO:0015881) |
| 0.1 | 1.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.9 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 1.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 1.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.8 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 3.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 2.7 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 3.4 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.8 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 | 0.4 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.5 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 1.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.9 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.9 | GO:0070055 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.1 | 0.5 | GO:1903764 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905024) |
| 0.1 | 0.9 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.3 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 0.6 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 2.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 2.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.3 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 1.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 1.7 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 1.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 1.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 4.0 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.1 | 0.7 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 2.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.4 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.5 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.6 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 1.0 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 2.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.3 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.1 | 1.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.3 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 5.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.7 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 2.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.6 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.4 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 1.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 1.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.3 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 1.0 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.8 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.4 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0098712 | L-glutamate import across plasma membrane(GO:0098712) |
| 0.0 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 1.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.9 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.0 | GO:0021679 | cerebellar molecular layer development(GO:0021679) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.3 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.3 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 1.1 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 2.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.2 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.6 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 1.4 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.6 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.9 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 2.0 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 1.5 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:0021924 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.5 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 2.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 2.4 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.3 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0071442 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.3 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.8 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.4 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.2 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.5 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.8 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.2 | 3.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.8 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 0.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 1.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 2.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 3.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.3 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 1.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 3.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.2 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.9 | 2.7 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.8 | 8.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 1.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.3 | 3.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 6.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.3 | 3.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.3 | 1.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.3 | 0.9 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.2 | 1.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 0.7 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.2 | 0.8 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.2 | 0.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 1.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.8 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 0.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 2.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 3.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 1.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 1.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 1.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 1.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 0.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.4 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 0.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 1.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.0 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 1.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.4 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) ethanol binding(GO:0035276) |
| 0.1 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 1.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 1.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.8 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 3.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 2.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 2.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 1.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 7.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 2.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.8 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.5 | GO:0019955 | cytokine binding(GO:0019955) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 4.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 4.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 6.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 3.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 2.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 2.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.3 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 4.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |