Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Mecom
Z-value: 0.75
Transcription factors associated with Mecom
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mecom
|
ENSMUSG00000027684.17 | MDS1 and EVI1 complex locus |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mecom | mm39_v1_chr3_-_30563919_30563971 | -0.59 | 1.5e-04 | Click! |
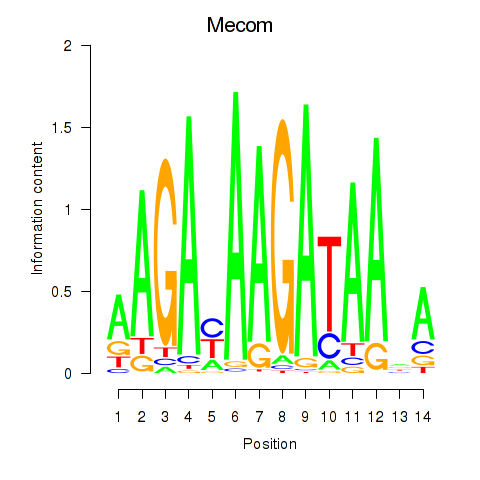
Activity profile of Mecom motif
Sorted Z-values of Mecom motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_62005498 | 21.13 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr4_-_61259801 | 19.89 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr6_+_138117295 | 7.48 |
ENSMUST00000008684.11
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr1_+_152275575 | 4.31 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr7_+_140415170 | 2.48 |
ENSMUST00000211372.2
ENSMUST00000026554.11 ENSMUST00000185612.3 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr7_+_140415431 | 2.24 |
ENSMUST00000209978.2
ENSMUST00000210916.2 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr4_+_132766793 | 2.18 |
ENSMUST00000105914.2
|
Ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr4_+_60003438 | 1.90 |
ENSMUST00000107517.8
ENSMUST00000107520.2 |
Mup6
|
major urinary protein 6 |
| chrX_+_21581135 | 1.69 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr10_+_61484331 | 1.49 |
ENSMUST00000020286.7
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr2_+_34764408 | 1.42 |
ENSMUST00000113068.9
ENSMUST00000047447.13 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr5_+_66417233 | 1.38 |
ENSMUST00000202994.4
ENSMUST00000201100.4 |
Nsun7
|
NOL1/NOP2/Sun domain family, member 7 |
| chr6_-_34887743 | 1.36 |
ENSMUST00000081214.12
|
Wdr91
|
WD repeat domain 91 |
| chr1_+_34275665 | 1.32 |
ENSMUST00000194192.3
|
Dst
|
dystonin |
| chr9_-_104214920 | 1.17 |
ENSMUST00000062723.14
ENSMUST00000215852.2 |
Acpp
|
acid phosphatase, prostate |
| chr5_-_136003294 | 1.15 |
ENSMUST00000154181.2
ENSMUST00000111152.8 ENSMUST00000111153.8 |
Ssc4d
|
scavenger receptor cysteine rich family, 4 domains |
| chr2_+_34764496 | 1.14 |
ENSMUST00000028228.6
|
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr15_-_96947963 | 1.11 |
ENSMUST00000230907.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr17_-_32639936 | 1.03 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr10_-_68114543 | 0.93 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr11_-_68277799 | 0.92 |
ENSMUST00000135141.2
|
Ntn1
|
netrin 1 |
| chr8_+_46944000 | 0.87 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr6_-_145156517 | 0.85 |
ENSMUST00000111728.8
ENSMUST00000204105.2 ENSMUST00000060797.10 |
Casc1
|
cancer susceptibility candidate 1 |
| chr11_-_115258493 | 0.83 |
ENSMUST00000123428.2
|
Hid1
|
HID1 domain containing |
| chrX_-_88453295 | 0.81 |
ENSMUST00000113959.8
ENSMUST00000113960.3 |
Dcaf8l
|
DDB1 and CUL4 associated factor 8 like |
| chrX_+_149981074 | 0.81 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr14_-_56448874 | 0.78 |
ENSMUST00000022757.5
|
Gzmf
|
granzyme F |
| chr16_-_48592319 | 0.78 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr1_-_189075515 | 0.76 |
ENSMUST00000193319.6
|
Kcnk2
|
potassium channel, subfamily K, member 2 |
| chr8_-_73188887 | 0.73 |
ENSMUST00000109974.2
|
Calr3
|
calreticulin 3 |
| chr11_-_69553390 | 0.66 |
ENSMUST00000129224.8
ENSMUST00000155200.8 |
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr16_-_48592372 | 0.65 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr9_-_104214899 | 0.64 |
ENSMUST00000112590.3
|
Acpp
|
acid phosphatase, prostate |
| chr11_-_69553451 | 0.63 |
ENSMUST00000018905.12
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr10_-_27492792 | 0.62 |
ENSMUST00000189575.2
|
Lama2
|
laminin, alpha 2 |
| chr2_-_45000250 | 0.62 |
ENSMUST00000201211.4
ENSMUST00000177302.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_30680502 | 0.62 |
ENSMUST00000133373.8
|
Cep41
|
centrosomal protein 41 |
| chr9_-_44437801 | 0.61 |
ENSMUST00000215661.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr13_+_19362068 | 0.61 |
ENSMUST00000103553.3
|
Trgv7
|
T cell receptor gamma, variable 7 |
| chr8_+_108669276 | 0.60 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr5_-_120605361 | 0.57 |
ENSMUST00000132916.2
|
Sdsl
|
serine dehydratase-like |
| chr1_+_119934624 | 0.56 |
ENSMUST00000072886.11
ENSMUST00000189037.2 |
Sctr
|
secretin receptor |
| chr5_+_34683141 | 0.55 |
ENSMUST00000125817.8
ENSMUST00000067638.14 |
Sh3bp2
|
SH3-domain binding protein 2 |
| chr9_+_59496571 | 0.54 |
ENSMUST00000121266.8
ENSMUST00000118164.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr18_+_84738996 | 0.54 |
ENSMUST00000235504.2
|
Dipk1c
|
divergent protein kinase domain 1C |
| chr10_+_63293284 | 0.52 |
ENSMUST00000105440.8
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr3_+_99161070 | 0.51 |
ENSMUST00000029462.10
|
Tbx15
|
T-box 15 |
| chr15_-_36140539 | 0.46 |
ENSMUST00000172831.8
|
Rgs22
|
regulator of G-protein signalling 22 |
| chr6_-_72343935 | 0.45 |
ENSMUST00000154098.3
|
Rnf181
|
ring finger protein 181 |
| chr10_+_3690348 | 0.44 |
ENSMUST00000120274.8
|
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chrX_+_11175063 | 0.44 |
ENSMUST00000178595.2
|
H2al1d
|
H2A histone family member L1D |
| chrX_-_112095181 | 0.44 |
ENSMUST00000026607.15
ENSMUST00000113388.3 |
Chm
|
choroidermia (RAB escort protein 1) |
| chr3_-_144511566 | 0.43 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr17_+_93506590 | 0.42 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr2_+_97298002 | 0.42 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr5_-_106606032 | 0.39 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chr1_-_162812087 | 0.39 |
ENSMUST00000028010.9
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr9_-_35523237 | 0.37 |
ENSMUST00000034610.4
|
Pate4
|
prostate and testis expressed 4 |
| chr5_+_65288418 | 0.36 |
ENSMUST00000101191.10
ENSMUST00000204348.3 |
Klhl5
|
kelch-like 5 |
| chr17_+_93506435 | 0.36 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr19_+_55886708 | 0.35 |
ENSMUST00000148666.3
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chrX_+_11187731 | 0.35 |
ENSMUST00000177926.3
|
H2al1h
|
H2A histone family member L1H |
| chr13_-_21476849 | 0.35 |
ENSMUST00000110491.9
|
Gpx5
|
glutathione peroxidase 5 |
| chr11_-_82910912 | 0.35 |
ENSMUST00000130822.3
|
Slfn8
|
schlafen 8 |
| chr8_-_43760017 | 0.34 |
ENSMUST00000082120.5
|
Zfp42
|
zinc finger protein 42 |
| chr4_+_65042411 | 0.33 |
ENSMUST00000084501.4
|
Pappa
|
pregnancy-associated plasma protein A |
| chr11_+_42312150 | 0.32 |
ENSMUST00000192403.2
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr14_-_51311892 | 0.32 |
ENSMUST00000216202.2
|
Olfr750
|
olfactory receptor 750 |
| chr9_+_72714156 | 0.31 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chrX_+_11178173 | 0.31 |
ENSMUST00000178979.2
|
H2al1e
|
H2A histone family member L1E |
| chr11_-_82911548 | 0.30 |
ENSMUST00000108152.9
|
Slfn8
|
schlafen 8 |
| chrX_+_11181397 | 0.30 |
ENSMUST00000179004.2
|
H2al1f
|
H2A histone family member L1F |
| chrX_+_11190898 | 0.30 |
ENSMUST00000164729.3
|
H2al1i
|
H2A histone family member L1I |
| chrX_+_11165496 | 0.27 |
ENSMUST00000188439.2
|
H2al1a
|
H2A histone family member L1A |
| chr17_-_37409147 | 0.27 |
ENSMUST00000216376.2
ENSMUST00000217372.2 |
Olfr91
|
olfactory receptor 91 |
| chrX_+_111513971 | 0.26 |
ENSMUST00000071814.13
|
Zfp711
|
zinc finger protein 711 |
| chr4_+_40920047 | 0.26 |
ENSMUST00000030122.5
|
Spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr11_-_50844572 | 0.26 |
ENSMUST00000162420.2
ENSMUST00000051159.3 |
Prop1
|
paired like homeodomain factor 1 |
| chr10_-_129948657 | 0.26 |
ENSMUST00000081469.2
|
Olfr823
|
olfactory receptor 823 |
| chr13_+_83672654 | 0.26 |
ENSMUST00000199019.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr17_-_37613523 | 0.25 |
ENSMUST00000215392.2
|
Olfr101
|
olfactory receptor 101 |
| chr6_+_24748324 | 0.25 |
ENSMUST00000031691.3
|
Hyal4
|
hyaluronoglucosaminidase 4 |
| chr6_+_113369377 | 0.25 |
ENSMUST00000032414.11
ENSMUST00000038889.12 |
Ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr3_+_41697046 | 0.24 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr17_+_33651864 | 0.23 |
ENSMUST00000174088.3
|
Actl9
|
actin-like 9 |
| chr7_-_102241093 | 0.23 |
ENSMUST00000213540.3
|
Olfr551
|
olfactory receptor 551 |
| chr11_-_82911615 | 0.23 |
ENSMUST00000038141.15
ENSMUST00000092838.11 |
Slfn8
|
schlafen 8 |
| chr16_-_57575070 | 0.22 |
ENSMUST00000089332.5
|
Col8a1
|
collagen, type VIII, alpha 1 |
| chr14_+_87654045 | 0.22 |
ENSMUST00000169504.8
ENSMUST00000168275.9 ENSMUST00000170865.8 |
Tdrd3
|
tudor domain containing 3 |
| chr11_-_59937302 | 0.22 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr13_-_58423461 | 0.22 |
ENSMUST00000223811.2
|
Gkap1
|
G kinase anchoring protein 1 |
| chr7_+_6225277 | 0.21 |
ENSMUST00000072662.12
ENSMUST00000155314.2 |
Zscan5b
|
zinc finger and SCAN domain containing 5B |
| chr12_-_11258973 | 0.20 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr17_-_33440626 | 0.20 |
ENSMUST00000213751.2
ENSMUST00000213642.2 ENSMUST00000215450.2 ENSMUST00000208645.3 |
Olfr1564
|
olfactory receptor 1564 |
| chrX_+_11184495 | 0.17 |
ENSMUST00000179859.2
|
H2al1g
|
H2A histone family member L1G |
| chr17_-_37430949 | 0.15 |
ENSMUST00000214994.2
ENSMUST00000216341.2 |
Olfr92
|
olfactory receptor 92 |
| chr16_-_29360301 | 0.14 |
ENSMUST00000057018.15
ENSMUST00000182627.8 |
Atp13a4
|
ATPase type 13A4 |
| chr16_+_94225942 | 0.12 |
ENSMUST00000141176.2
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr3_-_151953894 | 0.11 |
ENSMUST00000196529.5
|
Nexn
|
nexilin |
| chr13_-_27697185 | 0.11 |
ENSMUST00000018389.5
ENSMUST00000110350.9 |
Prl8a8
|
prolactin family 8, subfamily a, member 81 |
| chr9_+_39903409 | 0.11 |
ENSMUST00000217600.2
|
Olfr978
|
olfactory receptor 978 |
| chr2_-_125602234 | 0.10 |
ENSMUST00000139944.2
|
Secisbp2l
|
SECIS binding protein 2-like |
| chrX_+_47712676 | 0.09 |
ENSMUST00000177710.2
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr11_-_65636651 | 0.09 |
ENSMUST00000138093.2
|
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr12_-_35584968 | 0.09 |
ENSMUST00000116436.9
|
Ahr
|
aryl-hydrocarbon receptor |
| chr15_-_48655329 | 0.09 |
ENSMUST00000160658.8
ENSMUST00000100670.10 ENSMUST00000162830.8 |
Csmd3
|
CUB and Sushi multiple domains 3 |
| chr9_-_44437694 | 0.09 |
ENSMUST00000062215.8
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr3_-_123029745 | 0.09 |
ENSMUST00000106426.8
|
Synpo2
|
synaptopodin 2 |
| chrX_+_11171894 | 0.09 |
ENSMUST00000178806.2
|
H2al1c
|
H2A histone family member L1C |
| chr14_+_46616871 | 0.08 |
ENSMUST00000141358.2
|
Gm15217
|
predicted gene 15217 |
| chr3_-_123029782 | 0.08 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr2_-_90301592 | 0.08 |
ENSMUST00000111493.8
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chrX_-_153911405 | 0.08 |
ENSMUST00000076671.4
|
Cldn34b2
|
claudin 34B2 |
| chr19_-_37153436 | 0.07 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr6_+_65502344 | 0.06 |
ENSMUST00000212375.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chrX_+_9216866 | 0.06 |
ENSMUST00000178196.3
|
H2al1k
|
H2A histone family member L1K |
| chr5_-_151529486 | 0.05 |
ENSMUST00000233599.2
ENSMUST00000232905.2 |
Vmn2r18
|
vomeronasal 2, receptor 18 |
| chr6_+_96090127 | 0.05 |
ENSMUST00000122120.8
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr4_-_110149916 | 0.04 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr13_-_58422647 | 0.04 |
ENSMUST00000225034.2
|
Gkap1
|
G kinase anchoring protein 1 |
| chrX_+_11168671 | 0.03 |
ENSMUST00000189531.2
|
H2al1b
|
H2A histone family member L1B |
| chr9_+_38725910 | 0.03 |
ENSMUST00000213164.2
|
Olfr922
|
olfactory receptor 922 |
| chr14_+_34097422 | 0.03 |
ENSMUST00000111908.3
|
Mmrn2
|
multimerin 2 |
| chr10_-_107555840 | 0.02 |
ENSMUST00000050702.9
|
Ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chr9_-_102496047 | 0.02 |
ENSMUST00000215253.2
|
Cep63
|
centrosomal protein 63 |
| chr2_+_109522781 | 0.02 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr6_+_65502292 | 0.00 |
ENSMUST00000212402.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mecom
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.6 | 21.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.5 | 1.8 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.3 | 1.4 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.3 | 0.8 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.3 | 0.8 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 1.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 1.0 | GO:0032824 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.2 | 0.8 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.2 | 4.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.6 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.9 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.9 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 1.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.3 | GO:0060126 | hypophysis morphogenesis(GO:0048850) somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.3 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.5 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.4 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.3 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.3 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 2.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.7 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 0.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 8.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 1.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 4.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.6 | 1.8 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.6 | 1.7 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.4 | 1.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 7.5 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 21.1 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.2 | 0.6 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.2 | 0.9 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.2 | 1.0 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 4.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 0.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 2.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 0.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |