Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
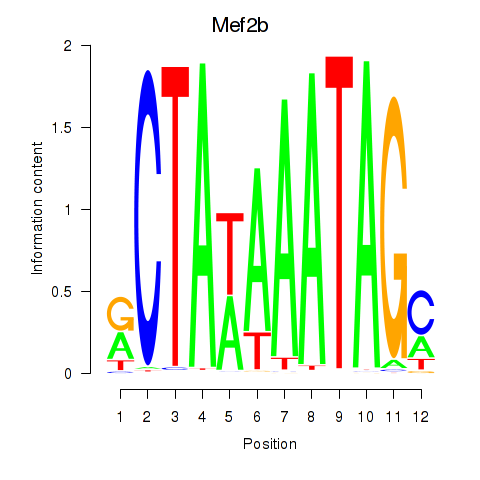
Results for Mef2b
Z-value: 0.69
Transcription factors associated with Mef2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2b
|
ENSMUSG00000079033.11 | myocyte enhancer factor 2B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2b | mm39_v1_chr8_+_70605404_70605431 | -0.32 | 6.1e-02 | Click! |
Activity profile of Mef2b motif
Sorted Z-values of Mef2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7779943 | 3.45 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr19_-_7780025 | 3.42 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr19_-_40175709 | 2.09 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr3_-_98721750 | 1.58 |
ENSMUST00000029463.13
|
Hsd3b6
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 6 |
| chr1_-_66974492 | 1.39 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr3_-_122828592 | 1.29 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr1_-_66974694 | 1.28 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr3_+_89366425 | 1.22 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr5_+_120614587 | 1.21 |
ENSMUST00000201684.4
ENSMUST00000066540.14 |
Sds
|
serine dehydratase |
| chr7_+_141995545 | 1.11 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr3_+_89366632 | 1.08 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase |
| chr7_+_141996067 | 1.07 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr10_-_88440869 | 1.01 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr7_-_48497771 | 1.00 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr14_-_20706556 | 0.98 |
ENSMUST00000090469.8
|
Myoz1
|
myozenin 1 |
| chr9_+_121606750 | 0.90 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40 |
| chr10_+_87694117 | 0.80 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr7_+_127603083 | 0.78 |
ENSMUST00000106248.8
|
Trim72
|
tripartite motif-containing 72 |
| chrX_+_156481906 | 0.78 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr15_+_25940912 | 0.78 |
ENSMUST00000226438.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr2_+_69500444 | 0.78 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr3_+_101993787 | 0.77 |
ENSMUST00000165540.9
ENSMUST00000164123.2 |
Casq2
|
calsequestrin 2 |
| chr11_+_67689094 | 0.77 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr3_+_101993731 | 0.76 |
ENSMUST00000029454.12
|
Casq2
|
calsequestrin 2 |
| chr7_+_44984681 | 0.75 |
ENSMUST00000085351.7
|
Hrc
|
histidine rich calcium binding protein |
| chr11_+_70548022 | 0.73 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr15_+_25940781 | 0.72 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr8_+_15107646 | 0.71 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr1_-_172047282 | 0.70 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chr5_+_122239007 | 0.69 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr5_+_122239030 | 0.68 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr17_-_45903494 | 0.66 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr5_-_116560916 | 0.66 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr7_-_127805518 | 0.64 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr2_-_6217844 | 0.62 |
ENSMUST00000042658.5
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr4_-_134099840 | 0.61 |
ENSMUST00000030643.3
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr7_+_80707328 | 0.61 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chr11_-_94867153 | 0.60 |
ENSMUST00000103162.8
ENSMUST00000166320.8 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr16_+_4825216 | 0.58 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr16_+_4825146 | 0.57 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr17_-_45903410 | 0.57 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr17_+_28451674 | 0.55 |
ENSMUST00000002320.16
ENSMUST00000232879.2 ENSMUST00000166744.8 |
Ppard
|
peroxisome proliferator activator receptor delta |
| chr11_+_70548513 | 0.53 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr16_+_4825170 | 0.52 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chrX_+_156482116 | 0.51 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr8_+_105688344 | 0.49 |
ENSMUST00000043183.8
|
Ces2g
|
carboxylesterase 2G |
| chr4_-_73869071 | 0.49 |
ENSMUST00000095023.2
ENSMUST00000030101.4 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr6_+_121277186 | 0.48 |
ENSMUST00000064580.14
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr17_-_45903188 | 0.47 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr15_+_25940931 | 0.46 |
ENSMUST00000110438.3
|
Retreg1
|
reticulophagy regulator 1 |
| chr3_-_86906591 | 0.45 |
ENSMUST00000063869.11
ENSMUST00000029717.4 |
Cd1d1
|
CD1d1 antigen |
| chr9_-_119852624 | 0.44 |
ENSMUST00000111635.4
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chrX_-_94521712 | 0.43 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr4_+_118965992 | 0.40 |
ENSMUST00000134105.8
ENSMUST00000144329.8 ENSMUST00000208090.2 |
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr1_+_155433858 | 0.39 |
ENSMUST00000080138.13
ENSMUST00000035560.9 ENSMUST00000097529.5 |
Acbd6
|
acyl-Coenzyme A binding domain containing 6 |
| chr2_-_160169414 | 0.38 |
ENSMUST00000099127.3
|
Gm826
|
predicted gene 826 |
| chr4_+_43493344 | 0.37 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr4_+_118965908 | 0.36 |
ENSMUST00000030398.10
|
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr5_-_24652775 | 0.35 |
ENSMUST00000123167.2
ENSMUST00000030799.15 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr1_-_75195127 | 0.34 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr3_-_158267771 | 0.33 |
ENSMUST00000199890.5
ENSMUST00000238317.3 ENSMUST00000200137.5 ENSMUST00000106044.6 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr17_+_12597490 | 0.33 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr15_+_25940859 | 0.33 |
ENSMUST00000226750.2
|
Retreg1
|
reticulophagy regulator 1 |
| chrX_+_163221035 | 0.33 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr13_+_89687915 | 0.32 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr3_+_32871669 | 0.32 |
ENSMUST00000072312.12
ENSMUST00000108228.8 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr11_+_77353431 | 0.31 |
ENSMUST00000130255.2
|
Coro6
|
coronin 6 |
| chrX_+_10583629 | 0.31 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr14_-_52151026 | 0.31 |
ENSMUST00000228164.2
|
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr5_+_115034997 | 0.31 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr7_-_103492361 | 0.30 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr18_+_67597929 | 0.30 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chrX_+_67805497 | 0.28 |
ENSMUST00000071848.7
|
Fmr1nb
|
Fmr1 neighbor |
| chr5_+_115373895 | 0.28 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr19_-_8700728 | 0.27 |
ENSMUST00000170157.8
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr11_-_54140462 | 0.27 |
ENSMUST00000019060.6
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr10_-_128384971 | 0.27 |
ENSMUST00000176906.2
|
Rpl41
|
ribosomal protein L41 |
| chr10_-_128384994 | 0.27 |
ENSMUST00000177163.8
ENSMUST00000176683.8 ENSMUST00000176010.8 |
Rpl41
|
ribosomal protein L41 |
| chr2_+_136555364 | 0.26 |
ENSMUST00000028727.11
ENSMUST00000110098.4 |
Snap25
|
synaptosomal-associated protein 25 |
| chr1_-_190915441 | 0.26 |
ENSMUST00000027941.14
|
Atf3
|
activating transcription factor 3 |
| chr5_-_28415020 | 0.25 |
ENSMUST00000118882.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr9_-_36708599 | 0.25 |
ENSMUST00000238932.2
ENSMUST00000115086.13 |
Ei24
|
etoposide induced 2.4 mRNA |
| chr2_+_90948481 | 0.25 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr10_-_95159410 | 0.24 |
ENSMUST00000217809.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr7_+_27878894 | 0.24 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr8_+_95584078 | 0.24 |
ENSMUST00000109521.4
|
Polr2c
|
polymerase (RNA) II (DNA directed) polypeptide C |
| chr9_-_36708569 | 0.24 |
ENSMUST00000163192.11
|
Ei24
|
etoposide induced 2.4 mRNA |
| chr14_-_20718337 | 0.23 |
ENSMUST00000057090.12
ENSMUST00000117386.2 |
Synpo2l
|
synaptopodin 2-like |
| chr6_-_88604404 | 0.23 |
ENSMUST00000120933.5
|
Kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr18_-_35631914 | 0.23 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr6_+_121277693 | 0.23 |
ENSMUST00000142419.2
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr15_+_98065039 | 0.23 |
ENSMUST00000031914.6
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr18_-_25887173 | 0.22 |
ENSMUST00000225477.2
|
Celf4
|
CUGBP, Elav-like family member 4 |
| chr5_-_28415166 | 0.22 |
ENSMUST00000117098.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr12_-_56583582 | 0.22 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr2_+_24076481 | 0.22 |
ENSMUST00000057567.3
|
Il1f9
|
interleukin 1 family, member 9 |
| chr2_-_57014015 | 0.22 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_-_14374794 | 0.21 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr6_+_48566540 | 0.21 |
ENSMUST00000204521.2
|
Gm45021
|
predicted gene 45021 |
| chr8_+_3550533 | 0.21 |
ENSMUST00000208306.2
|
Mcoln1
|
mucolipin 1 |
| chr14_+_55813074 | 0.21 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr15_+_76211597 | 0.21 |
ENSMUST00000059045.8
|
Exosc4
|
exosome component 4 |
| chr1_-_14374842 | 0.20 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr14_-_52151537 | 0.20 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr10_-_95158827 | 0.20 |
ENSMUST00000220279.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chrX_+_67805443 | 0.20 |
ENSMUST00000069731.12
ENSMUST00000114647.8 |
Fmr1nb
|
Fmr1 neighbor |
| chr16_-_93400691 | 0.20 |
ENSMUST00000023669.14
ENSMUST00000233931.2 ENSMUST00000154355.3 |
Setd4
|
SET domain containing 4 |
| chr7_-_142453722 | 0.19 |
ENSMUST00000000219.10
|
Th
|
tyrosine hydroxylase |
| chr10_-_18619658 | 0.19 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr18_+_76192529 | 0.19 |
ENSMUST00000167921.2
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr18_-_3280999 | 0.19 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chrX_-_63320543 | 0.19 |
ENSMUST00000114679.2
ENSMUST00000069926.14 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr11_-_79394904 | 0.18 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr7_-_105230479 | 0.18 |
ENSMUST00000191601.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr7_+_27879650 | 0.18 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr1_+_160806194 | 0.18 |
ENSMUST00000064725.11
ENSMUST00000191936.2 |
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr11_-_74228857 | 0.18 |
ENSMUST00000214490.2
|
Olfr410
|
olfactory receptor 410 |
| chr13_-_24118139 | 0.18 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr10_+_32959472 | 0.18 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr13_-_23882437 | 0.18 |
ENSMUST00000102967.3
|
H4c3
|
H4 clustered histone 3 |
| chr19_+_6144449 | 0.18 |
ENSMUST00000235513.2
|
Gm550
|
predicted gene 550 |
| chr8_+_120588977 | 0.18 |
ENSMUST00000034287.10
|
Klhl36
|
kelch-like 36 |
| chr6_+_137387718 | 0.18 |
ENSMUST00000167002.4
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr11_-_74228504 | 0.18 |
ENSMUST00000213831.2
|
Olfr410
|
olfactory receptor 410 |
| chr18_+_61096597 | 0.18 |
ENSMUST00000115295.9
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr5_+_92534738 | 0.18 |
ENSMUST00000128246.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr1_+_160806241 | 0.17 |
ENSMUST00000195760.2
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr8_+_95584146 | 0.17 |
ENSMUST00000211939.2
ENSMUST00000212124.2 |
Polr2c
|
polymerase (RNA) II (DNA directed) polypeptide C |
| chr7_+_81220987 | 0.17 |
ENSMUST00000165460.2
|
Whamm
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr11_+_54195006 | 0.17 |
ENSMUST00000108904.10
ENSMUST00000108905.10 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr18_+_37093321 | 0.17 |
ENSMUST00000192168.2
|
Pcdha5
|
protocadherin alpha 5 |
| chr5_-_134935579 | 0.17 |
ENSMUST00000201316.4
ENSMUST00000047305.6 |
Tmem270
|
transmembrane protein 270 |
| chr2_-_30068433 | 0.17 |
ENSMUST00000100220.5
|
Spout1
|
SPOUT domain containing methyltransferase 1 |
| chr7_-_101749433 | 0.17 |
ENSMUST00000106937.8
|
Art5
|
ADP-ribosyltransferase 5 |
| chr7_+_101546059 | 0.17 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chrX_-_88453295 | 0.17 |
ENSMUST00000113959.8
ENSMUST00000113960.3 |
Dcaf8l
|
DDB1 and CUL4 associated factor 8 like |
| chr9_-_54554483 | 0.16 |
ENSMUST00000128163.8
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr15_-_102630589 | 0.16 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr2_+_111205867 | 0.16 |
ENSMUST00000062407.6
|
Olfr1284
|
olfactory receptor 1284 |
| chr2_+_164328375 | 0.16 |
ENSMUST00000069385.15
ENSMUST00000143690.8 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr7_+_15863679 | 0.16 |
ENSMUST00000211649.2
|
Slc8a2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr14_-_36832044 | 0.16 |
ENSMUST00000179488.3
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr6_-_68857658 | 0.16 |
ENSMUST00000198756.2
|
Gm42543
|
predicted gene 42543 |
| chr2_+_31204314 | 0.16 |
ENSMUST00000113532.9
|
Hmcn2
|
hemicentin 2 |
| chr11_+_54194831 | 0.15 |
ENSMUST00000000145.12
ENSMUST00000138515.8 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr1_+_15382676 | 0.15 |
ENSMUST00000170146.3
|
Kcnb2
|
potassium voltage gated channel, Shab-related subfamily, member 2 |
| chr11_+_54194624 | 0.15 |
ENSMUST00000093106.12
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr4_+_43851565 | 0.15 |
ENSMUST00000107860.3
|
Olfr155
|
olfactory receptor 155 |
| chr14_+_53743184 | 0.15 |
ENSMUST00000103583.5
|
Trav10
|
T cell receptor alpha variable 10 |
| chr1_+_78794475 | 0.15 |
ENSMUST00000057262.8
ENSMUST00000187432.2 |
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
| chr16_-_17019352 | 0.15 |
ENSMUST00000090192.12
|
Ube2l3
|
ubiquitin-conjugating enzyme E2L 3 |
| chr7_+_143420586 | 0.15 |
ENSMUST00000152910.3
ENSMUST00000207630.2 |
Acte1
|
actin, epsilon 1 |
| chr8_+_3550450 | 0.14 |
ENSMUST00000004683.13
ENSMUST00000160338.2 |
Mcoln1
|
mucolipin 1 |
| chrX_-_72442342 | 0.14 |
ENSMUST00000180787.3
|
Gm18336
|
predicted gene, 18336 |
| chr3_-_154760978 | 0.14 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr2_-_87838612 | 0.14 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr3_-_64049349 | 0.14 |
ENSMUST00000177151.9
|
Vmn2r2
|
vomeronasal 2, receptor 2 |
| chr7_-_104570689 | 0.14 |
ENSMUST00000217177.2
|
Olfr667
|
olfactory receptor 667 |
| chr9_+_102382949 | 0.14 |
ENSMUST00000039390.6
|
Ky
|
kyphoscoliosis peptidase |
| chr6_+_137387729 | 0.14 |
ENSMUST00000203914.2
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr17_-_18498018 | 0.14 |
ENSMUST00000172190.4
ENSMUST00000231815.3 |
Vmn2r94
|
vomeronasal 2, receptor 94 |
| chr9_-_55917834 | 0.14 |
ENSMUST00000217105.2
|
4930563M21Rik
|
RIKEN cDNA 4930563M21 gene |
| chr17_+_44337566 | 0.14 |
ENSMUST00000229939.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr4_+_156300325 | 0.13 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr11_-_53509485 | 0.13 |
ENSMUST00000000889.7
|
Il4
|
interleukin 4 |
| chr12_-_40249489 | 0.13 |
ENSMUST00000220951.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr5_+_140404997 | 0.13 |
ENSMUST00000100507.8
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
| chr14_-_51134930 | 0.13 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr7_-_105230395 | 0.13 |
ENSMUST00000188726.2
ENSMUST00000188440.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr3_-_123029782 | 0.13 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr18_-_3281089 | 0.13 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr19_-_11291805 | 0.13 |
ENSMUST00000187467.2
|
Ms4a14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr13_+_83672389 | 0.13 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr2_+_80122811 | 0.12 |
ENSMUST00000057072.6
|
Prdx6b
|
peroxiredoxin 6B |
| chr5_-_143963413 | 0.12 |
ENSMUST00000031622.13
|
Ocm
|
oncomodulin |
| chr18_+_66006119 | 0.12 |
ENSMUST00000025395.10
|
Grp
|
gastrin releasing peptide |
| chr8_+_20478993 | 0.12 |
ENSMUST00000211848.2
|
Gm31371
|
predicted gene, 31371 |
| chr7_+_99384352 | 0.12 |
ENSMUST00000098264.2
|
Olfr520
|
olfactory receptor 520 |
| chr1_-_4430481 | 0.12 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr10_-_21943978 | 0.12 |
ENSMUST00000092672.6
|
4930444G20Rik
|
RIKEN cDNA 4930444G20 gene |
| chr2_+_79538124 | 0.12 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr15_+_94527117 | 0.12 |
ENSMUST00000080141.6
|
Tmem117
|
transmembrane protein 117 |
| chr7_-_25374472 | 0.12 |
ENSMUST00000108404.8
ENSMUST00000108405.2 ENSMUST00000079439.10 |
Tmem91
|
transmembrane protein 91 |
| chr1_-_39844467 | 0.12 |
ENSMUST00000171319.4
|
Gm3646
|
predicted gene 3646 |
| chr5_+_73164226 | 0.11 |
ENSMUST00000031127.11
ENSMUST00000201304.2 |
Slc10a4
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 4 |
| chr18_+_37646674 | 0.11 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
| chr6_-_55109960 | 0.11 |
ENSMUST00000003568.15
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr2_-_87798643 | 0.11 |
ENSMUST00000099841.4
|
Olfr1157
|
olfactory receptor 1157 |
| chr18_-_3299536 | 0.11 |
ENSMUST00000129435.8
ENSMUST00000122958.8 |
Crem
|
cAMP responsive element modulator |
| chr11_-_101315345 | 0.11 |
ENSMUST00000107257.8
ENSMUST00000107259.4 ENSMUST00000107252.9 ENSMUST00000093933.11 |
Gm27029
Ptges3l
|
predicted gene, 27029 prostaglandin E synthase 3 like |
| chr9_-_19799300 | 0.11 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862 |
| chr18_+_66005891 | 0.11 |
ENSMUST00000173985.10
|
Grp
|
gastrin releasing peptide |
| chr2_+_164328763 | 0.11 |
ENSMUST00000109349.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr14_+_26616514 | 0.11 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr2_-_79738734 | 0.11 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr8_-_106670014 | 0.11 |
ENSMUST00000038896.8
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr17_-_20268598 | 0.11 |
ENSMUST00000168050.3
|
Vmn2r104
|
vomeronasal 2, receptor 104 |
| chr8_-_13888389 | 0.11 |
ENSMUST00000071308.6
|
AF366264
|
cDNA sequence AF366264 |
| chr13_-_21826688 | 0.10 |
ENSMUST00000205631.3
|
Olfr11
|
olfactory receptor 11 |
| chr7_+_144415115 | 0.10 |
ENSMUST00000060336.5
|
Fgf4
|
fibroblast growth factor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.5 | 6.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.5 | 1.4 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.3 | 1.0 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.3 | 2.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 1.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.7 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.2 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 1.5 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.2 | 0.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.5 | GO:0048007 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 1.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.8 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.9 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.5 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.3 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.5 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 1.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.1 | 0.3 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.1 | 0.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.3 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.3 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 1.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.5 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:1900738 | psychomotor behavior(GO:0036343) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 2.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.0 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.0 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 2.0 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 4.3 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.3 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.0 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 0.5 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 1.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 0.8 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.2 | 1.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.2 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.1 | 1.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.3 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.6 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 4.4 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 1.2 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.4 | 6.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 2.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 0.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 0.6 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.2 | 0.8 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 1.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 1.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 2.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 2.3 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 1.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |