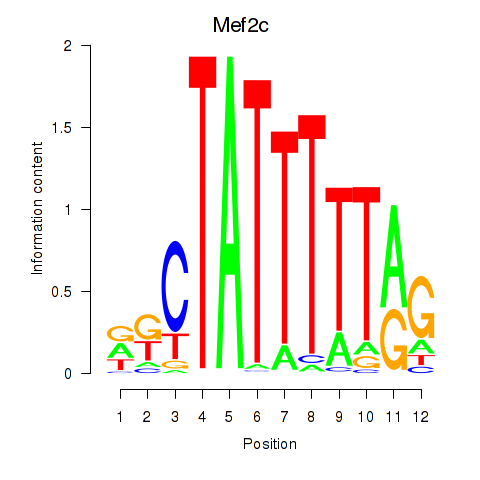
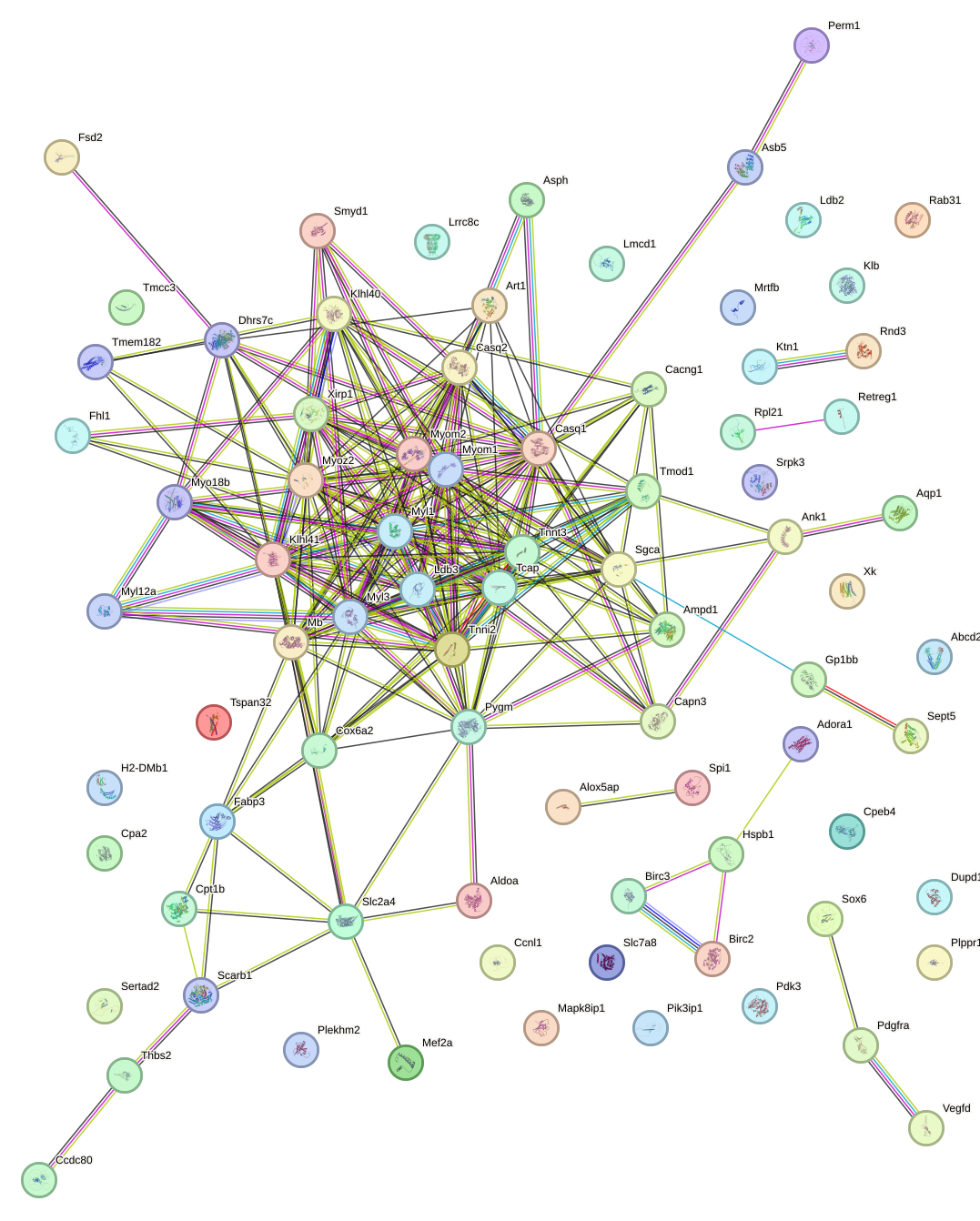
|
chr7_+_141995545
Show fit
|
9.36 |
ENSMUST00000105971.8
ENSMUST00000145287.8
|
Tnni2
|
troponin I, skeletal, fast 2
|
|
chr1_-_66974492
Show fit
|
6.25 |
ENSMUST00000120415.8
ENSMUST00000119429.8
|
Myl1
|
myosin, light polypeptide 1
|
|
chr7_+_142052569
Show fit
|
6.19 |
ENSMUST00000078497.15
ENSMUST00000105953.10
ENSMUST00000179658.8
ENSMUST00000105954.10
ENSMUST00000105952.10
ENSMUST00000105955.8
ENSMUST00000074187.13
ENSMUST00000169299.9
ENSMUST00000105957.10
ENSMUST00000180152.8
ENSMUST00000105950.11
ENSMUST00000105958.10
ENSMUST00000105949.8
|
Tnnt3
|
troponin T3, skeletal, fast
|
|
chr7_+_141996067
Show fit
|
5.33 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2
|
|
chr1_-_66974694
Show fit
|
5.15 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1
|
|
chr11_+_98274637
Show fit
|
5.03 |
ENSMUST00000008021.3
|
Tcap
|
titin-cap
|
|
chr1_-_172047282
Show fit
|
4.15 |
ENSMUST00000170700.2
ENSMUST00000003554.11
|
Casq1
|
calsequestrin 1
|
|
chr7_-_127805518
Show fit
|
4.05 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2
|
|
chr5_+_122239007
Show fit
|
3.98 |
ENSMUST00000014080.13
ENSMUST00000111750.8
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow
|
|
chr3_-_122828592
Show fit
|
3.95 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2
|
|
chr11_+_70548022
Show fit
|
3.86 |
ENSMUST00000157027.8
ENSMUST00000072841.12
ENSMUST00000108548.8
ENSMUST00000126241.8
|
Eno3
|
enolase 3, beta muscle
|
|
chr5_+_122239030
Show fit
|
3.55 |
ENSMUST00000139213.8
ENSMUST00000111751.8
ENSMUST00000155612.8
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow
|
|
chr10_-_88440869
Show fit
|
3.40 |
ENSMUST00000119185.8
ENSMUST00000238199.2
|
Mybpc1
|
myosin binding protein C, slow-type
|
|
chr19_+_6434416
Show fit
|
3.13 |
ENSMUST00000035269.15
ENSMUST00000113483.2
|
Pygm
|
muscle glycogen phosphorylase
|
|
chr4_+_130202388
Show fit
|
2.88 |
ENSMUST00000070532.8
|
Fabp3
|
fatty acid binding protein 3, muscle and heart
|
|
chr10_-_88440996
Show fit
|
2.78 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type
|
|
chr11_+_120839879
Show fit
|
2.53 |
ENSMUST00000154187.8
ENSMUST00000100130.10
ENSMUST00000129473.8
ENSMUST00000168579.8
|
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3
|
|
chr3_+_101993787
Show fit
|
2.50 |
ENSMUST00000165540.9
ENSMUST00000164123.2
|
Casq2
|
calsequestrin 2
|
|
chr4_+_46039202
Show fit
|
2.32 |
ENSMUST00000156200.8
|
Tmod1
|
tropomodulin 1
|
|
chr14_-_34310602
Show fit
|
2.31 |
ENSMUST00000064098.14
ENSMUST00000090040.12
ENSMUST00000022330.9
ENSMUST00000022327.13
|
Ldb3
|
LIM domain binding 3
|
|
chr1_+_135727140
Show fit
|
2.25 |
ENSMUST00000152208.8
ENSMUST00000152075.8
|
Tnni1
|
troponin I, skeletal, slow 1
|
|
chr17_+_71326510
Show fit
|
2.20 |
ENSMUST00000073211.13
ENSMUST00000024847.14
|
Myom1
|
myomesin 1
|
|
chr11_+_70548513
Show fit
|
2.13 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle
|
|
chr3_+_101993731
Show fit
|
2.05 |
ENSMUST00000029454.12
|
Casq2
|
calsequestrin 2
|
|
chr9_+_110592709
Show fit
|
2.03 |
ENSMUST00000079784.12
|
Myl3
|
myosin, light polypeptide 3
|
|
chr14_-_34310637
Show fit
|
1.99 |
ENSMUST00000227819.2
|
Ldb3
|
LIM domain binding 3
|
|
chr6_+_112250719
Show fit
|
1.98 |
ENSMUST00000032376.6
|
Lmcd1
|
LIM and cysteine-rich domains 1
|
|
chr15_-_76906832
Show fit
|
1.95 |
ENSMUST00000019037.10
ENSMUST00000169226.9
|
Mb
|
myoglobin
|
|
chr11_+_67689094
Show fit
|
1.95 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr15_-_89310060
Show fit
|
1.80 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle
|
|
chr6_+_30541581
Show fit
|
1.70 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic
|
|
chr7_+_101750943
Show fit
|
1.69 |
ENSMUST00000033300.4
|
Art1
|
ADP-ribosyltransferase 1
|
|
chr15_-_91075933
Show fit
|
1.68 |
ENSMUST00000069511.8
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2
|
|
chr16_-_18440388
Show fit
|
1.66 |
ENSMUST00000167388.3
|
Gp1bb
|
glycoprotein Ib, beta polypeptide
|
|
chr1_+_135727228
Show fit
|
1.64 |
ENSMUST00000154463.8
ENSMUST00000139986.8
|
Tnni1
|
troponin I, skeletal, slow 1
|
|
chr5_-_18093739
Show fit
|
1.62 |
ENSMUST00000169095.6
ENSMUST00000197574.2
|
Cd36
|
CD36 molecule
|
|
chr5_+_135916764
Show fit
|
1.57 |
ENSMUST00000005077.7
|
Hspb1
|
heat shock protein 1
|
|
chr8_+_15107646
Show fit
|
1.57 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2
|
|
chr11_-_107607343
Show fit
|
1.55 |
ENSMUST00000021065.6
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1
|
|
chr17_+_71326542
Show fit
|
1.54 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1
|
|
chrX_+_55825033
Show fit
|
1.49 |
ENSMUST00000114772.9
ENSMUST00000114768.10
ENSMUST00000155882.8
|
Fhl1
|
four and a half LIM domains 1
|
|
chr5_+_135916847
Show fit
|
1.47 |
ENSMUST00000111155.2
|
Hspb1
|
heat shock protein 1
|
|
chr2_+_69500444
Show fit
|
1.42 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41
|
|
chr1_+_40844739
Show fit
|
1.38 |
ENSMUST00000114765.4
|
Tmem182
|
transmembrane protein 182
|
|
chr1_+_135727571
Show fit
|
1.38 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1
|
|
chr7_+_127603083
Show fit
|
1.38 |
ENSMUST00000106248.8
|
Trim72
|
tripartite motif-containing 72
|
|
chr11_-_69838971
Show fit
|
1.34 |
ENSMUST00000179298.3
ENSMUST00000018710.13
ENSMUST00000135437.3
ENSMUST00000141837.9
ENSMUST00000142500.8
|
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr15_-_78090591
Show fit
|
1.33 |
ENSMUST00000120592.2
|
Pvalb
|
parvalbumin
|
|
chr5_-_113044216
Show fit
|
1.33 |
ENSMUST00000086617.11
|
Myo18b
|
myosin XVIIIb
|
|
chr15_+_73620213
Show fit
|
1.31 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3
|
|
chr11_-_94867153
Show fit
|
1.28 |
ENSMUST00000103162.8
ENSMUST00000166320.8
|
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein)
|
|
chrX_+_55824797
Show fit
|
1.27 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1
|
|
chr7_-_4517367
Show fit
|
1.26 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow
|
|
chrX_-_92875712
Show fit
|
1.26 |
ENSMUST00000045748.7
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3
|
|
chr3_-_142101339
Show fit
|
1.26 |
ENSMUST00000198381.5
ENSMUST00000090134.12
ENSMUST00000196908.5
|
Pdlim5
|
PDZ and LIM domain 5
|
|
chr15_+_25940931
Show fit
|
1.20 |
ENSMUST00000110438.3
|
Retreg1
|
reticulophagy regulator 1
|
|
chr4_+_49059255
Show fit
|
1.20 |
ENSMUST00000076670.3
|
Plppr1
|
phospholipid phosphatase related 1
|
|
chr5_+_149202157
Show fit
|
1.13 |
ENSMUST00000200806.4
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein
|
|
chr7_-_126399208
Show fit
|
1.11 |
ENSMUST00000133514.8
ENSMUST00000151137.8
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr9_+_121606750
Show fit
|
1.10 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40
|
|
chr17_-_14914484
Show fit
|
1.06 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2
|
|
chr3_+_102981326
Show fit
|
1.05 |
ENSMUST00000090715.13
ENSMUST00000155034.6
|
Ampd1
|
adenosine monophosphate deaminase 1
|
|
chr9_-_119852624
Show fit
|
1.05 |
ENSMUST00000111635.4
|
Xirp1
|
xin actin-binding repeat containing 1
|
|
chr7_-_4517559
Show fit
|
1.03 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow
|
|
chr7_+_130467564
Show fit
|
1.02 |
ENSMUST00000075181.11
ENSMUST00000151119.9
ENSMUST00000048180.12
|
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr3_-_65865656
Show fit
|
1.02 |
ENSMUST00000029416.14
|
Ccnl1
|
cyclin L1
|
|
chr7_-_126399778
Show fit
|
1.00 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr7_-_4517608
Show fit
|
0.98 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow
|
|
chr15_+_25940781
Show fit
|
0.97 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1
|
|
chr1_+_135764092
Show fit
|
0.92 |
ENSMUST00000188028.7
ENSMUST00000178204.8
ENSMUST00000190451.7
ENSMUST00000189732.7
ENSMUST00000189355.7
|
Tnnt2
|
troponin T2, cardiac
|
|
chr15_+_25940859
Show fit
|
0.90 |
ENSMUST00000226750.2
|
Retreg1
|
reticulophagy regulator 1
|
|
chr6_-_142647985
Show fit
|
0.88 |
ENSMUST00000205202.3
ENSMUST00000073173.12
ENSMUST00000111771.8
|
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr14_-_20718337
Show fit
|
0.87 |
ENSMUST00000057090.12
ENSMUST00000117386.2
|
Synpo2l
|
synaptopodin 2-like
|
|
chr5_+_105667254
Show fit
|
0.87 |
ENSMUST00000067924.13
ENSMUST00000150981.2
|
Lrrc8c
|
leucine rich repeat containing 8 family, member C
|
|
chr7_-_126399574
Show fit
|
0.87 |
ENSMUST00000106348.8
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr15_+_25940912
Show fit
|
0.86 |
ENSMUST00000226438.2
|
Retreg1
|
reticulophagy regulator 1
|
|
chr14_-_34310438
Show fit
|
0.85 |
ENSMUST00000228044.2
ENSMUST00000022328.14
|
Ldb3
|
LIM domain binding 3
|
|
chr14_+_47886748
Show fit
|
0.85 |
ENSMUST00000190252.7
ENSMUST00000186466.3
|
Ktn1
|
kinectin 1
|
|
chr11_+_3280401
Show fit
|
0.84 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr7_+_130467486
Show fit
|
0.83 |
ENSMUST00000120441.8
|
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr14_-_55019387
Show fit
|
0.82 |
ENSMUST00000022787.8
|
Slc7a8
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 8
|
|
chr15_+_102391614
Show fit
|
0.81 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2
|
|
chrX_+_72818003
Show fit
|
0.81 |
ENSMUST00000002081.6
|
Srpk3
|
serine/arginine-rich protein specific kinase 3
|
|
chrX_+_9138995
Show fit
|
0.79 |
ENSMUST00000015486.7
|
Xk
|
X-linked Kx blood group
|
|
chr2_+_90927053
Show fit
|
0.77 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene
|
|
chr7_-_67022520
Show fit
|
0.77 |
ENSMUST00000156690.8
ENSMUST00000107476.8
ENSMUST00000076325.12
ENSMUST00000032776.15
ENSMUST00000133074.2
|
Mef2a
|
myocyte enhancer factor 2A
|
|
chr4_+_156300325
Show fit
|
0.76 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1
|
|
chr8_+_23548541
Show fit
|
0.73 |
ENSMUST00000173248.8
|
Ank1
|
ankyrin 1, erythroid
|
|
chr7_+_46495521
Show fit
|
0.68 |
ENSMUST00000133062.2
|
Ldha
|
lactate dehydrogenase A
|
|
chr6_-_142647944
Show fit
|
0.65 |
ENSMUST00000100827.5
ENSMUST00000087527.11
|
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr4_-_9643636
Show fit
|
0.64 |
ENSMUST00000108333.8
ENSMUST00000108334.8
ENSMUST00000108335.8
ENSMUST00000152526.8
ENSMUST00000103004.10
|
Asph
|
aspartate-beta-hydroxylase
|
|
chr10_+_94386714
Show fit
|
0.61 |
ENSMUST00000148910.3
ENSMUST00000117460.8
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr11_+_31822211
Show fit
|
0.60 |
ENSMUST00000020543.13
ENSMUST00000109412.9
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr16_+_44913974
Show fit
|
0.59 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr5_+_65505657
Show fit
|
0.59 |
ENSMUST00000031096.11
|
Klb
|
klotho beta
|
|
chr6_-_70051586
Show fit
|
0.59 |
ENSMUST00000103377.3
|
Igkv6-32
|
immunoglobulin kappa variable 6-32
|
|
chr17_-_66079681
Show fit
|
0.59 |
ENSMUST00000070673.9
|
Rab31
|
RAB31, member RAS oncogene family
|
|
chr7_-_81216687
Show fit
|
0.57 |
ENSMUST00000042318.6
|
Fsd2
|
fibronectin type III and SPRY domain containing 2
|
|
chr5_+_75312939
Show fit
|
0.56 |
ENSMUST00000202681.4
ENSMUST00000000476.15
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide
|
|
chr2_+_120294046
Show fit
|
0.55 |
ENSMUST00000028749.15
ENSMUST00000110721.9
ENSMUST00000239364.2
|
Capn3
|
calpain 3
|
|
chr6_-_71239216
Show fit
|
0.55 |
ENSMUST00000129630.3
ENSMUST00000114186.9
ENSMUST00000074301.10
|
Smyd1
|
SET and MYND domain containing 1
|
|
chr5_+_146769700
Show fit
|
0.54 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21
|
|
chr2_-_92222979
Show fit
|
0.53 |
ENSMUST00000111279.9
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chrX_+_163156359
Show fit
|
0.52 |
ENSMUST00000033751.8
|
Vegfd
|
vascular endothelial growth factor D
|
|
chr4_-_141391406
Show fit
|
0.49 |
ENSMUST00000084203.11
|
Plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2
|
|
chr3_-_20209260
Show fit
|
0.48 |
ENSMUST00000178328.8
|
Gyg
|
glycogenin
|
|
chr7_-_115630282
Show fit
|
0.47 |
ENSMUST00000206034.2
ENSMUST00000106612.8
|
Sox6
|
SRY (sex determining region Y)-box 6
|
|
chr14_-_20714634
Show fit
|
0.47 |
ENSMUST00000119483.2
|
Synpo2l
|
synaptopodin 2-like
|
|
chr3_-_20209220
Show fit
|
0.47 |
ENSMUST00000184552.2
|
Gyg
|
glycogenin
|
|
chr17_-_71305003
Show fit
|
0.47 |
ENSMUST00000024846.13
ENSMUST00000232766.2
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr8_-_41469786
Show fit
|
0.46 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1
|
|
chr13_+_38010879
Show fit
|
0.46 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1
|
|
chr11_+_20597149
Show fit
|
0.45 |
ENSMUST00000109585.2
|
Sertad2
|
SERTA domain containing 2
|
|
chr1_-_134162231
Show fit
|
0.44 |
ENSMUST00000169927.2
|
Adora1
|
adenosine A1 receptor
|
|
chr7_-_115445315
Show fit
|
0.43 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6
|
|
chr8_+_55024446
Show fit
|
0.43 |
ENSMUST00000239166.2
ENSMUST00000239106.2
ENSMUST00000239152.2
|
Asb5
|
ankyrin repeat and SOCs box-containing 5
|
|
chr14_-_21764638
Show fit
|
0.42 |
ENSMUST00000073870.7
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1
|
|
chr2_-_51039112
Show fit
|
0.41 |
ENSMUST00000154545.2
ENSMUST00000017288.9
|
Rnd3
|
Rho family GTPase 3
|
|
chr16_+_13176238
Show fit
|
0.40 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B
|
|
chr2_+_71884943
Show fit
|
0.40 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr6_+_55313409
Show fit
|
0.40 |
ENSMUST00000004774.4
|
Aqp1
|
aquaporin 1
|
|
chr9_-_7873171
Show fit
|
0.39 |
ENSMUST00000159323.2
ENSMUST00000115673.3
|
Birc3
|
baculoviral IAP repeat-containing 3
|
|
chr13_+_5911481
Show fit
|
0.39 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6
|
|
chr11_+_82802079
Show fit
|
0.38 |
ENSMUST00000018989.14
ENSMUST00000164945.3
|
Unc45b
|
unc-45 myosin chaperone B
|
|
chr10_-_61814852
Show fit
|
0.38 |
ENSMUST00000105453.8
ENSMUST00000105452.9
ENSMUST00000105454.3
|
Col13a1
|
collagen, type XIII, alpha 1
|
|
chr4_+_137589581
Show fit
|
0.37 |
ENSMUST00000130407.3
|
Ece1
|
endothelin converting enzyme 1
|
|
chr4_-_73869071
Show fit
|
0.37 |
ENSMUST00000095023.2
ENSMUST00000030101.4
|
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene
|
|
chr19_-_40365318
Show fit
|
0.37 |
ENSMUST00000239304.2
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr13_+_38388904
Show fit
|
0.37 |
ENSMUST00000091641.13
ENSMUST00000178564.2
|
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12)
|
|
chr6_-_128503666
Show fit
|
0.36 |
ENSMUST00000143664.2
ENSMUST00000112132.8
|
Pzp
|
PZP, alpha-2-macroglobulin like
|
|
chr2_-_76812799
Show fit
|
0.36 |
ENSMUST00000011934.13
ENSMUST00000099981.10
ENSMUST00000099980.10
ENSMUST00000111882.9
ENSMUST00000140091.8
|
Ttn
|
titin
|
|
chr10_-_8638215
Show fit
|
0.36 |
ENSMUST00000212553.2
|
Sash1
|
SAM and SH3 domain containing 1
|
|
chr17_+_57556449
Show fit
|
0.36 |
ENSMUST00000224947.2
ENSMUST00000019631.11
ENSMUST00000224885.2
ENSMUST00000224152.2
|
Trip10
|
thyroid hormone receptor interactor 10
|
|
chr12_+_117621644
Show fit
|
0.36 |
ENSMUST00000222105.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr13_+_30529498
Show fit
|
0.35 |
ENSMUST00000221743.2
|
Agtr1a
|
angiotensin II receptor, type 1a
|
|
chr15_+_102379503
Show fit
|
0.35 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2
|
|
chr11_+_23234644
Show fit
|
0.35 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1
|
|
chr14_-_32407203
Show fit
|
0.34 |
ENSMUST00000096038.4
|
3425401B19Rik
|
RIKEN cDNA 3425401B19 gene
|
|
chr11_-_59029996
Show fit
|
0.34 |
ENSMUST00000219084.3
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF
|
|
chr16_-_56533179
Show fit
|
0.33 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene
|
|
chr11_+_67128843
Show fit
|
0.32 |
ENSMUST00000018632.11
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle
|
|
chr12_+_59177780
Show fit
|
0.32 |
ENSMUST00000177460.8
|
Mia2
|
MIA SH3 domain ER export factor 2
|
|
chr12_-_103304573
Show fit
|
0.32 |
ENSMUST00000149431.2
|
Asb2
|
ankyrin repeat and SOCS box-containing 2
|
|
chr3_-_142101418
Show fit
|
0.31 |
ENSMUST00000029941.16
ENSMUST00000058626.9
|
Pdlim5
|
PDZ and LIM domain 5
|
|
chr15_+_80507671
Show fit
|
0.30 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2
|
|
chr13_+_38010203
Show fit
|
0.29 |
ENSMUST00000128570.9
|
Rreb1
|
ras responsive element binding protein 1
|
|
chrX_-_94521712
Show fit
|
0.29 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12
|
|
chr9_-_7873017
Show fit
|
0.29 |
ENSMUST00000013949.15
|
Birc3
|
baculoviral IAP repeat-containing 3
|
|
chr6_+_108190050
Show fit
|
0.29 |
ENSMUST00000032192.9
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1
|
|
chr13_+_102830029
Show fit
|
0.29 |
ENSMUST00000022124.10
ENSMUST00000171267.2
ENSMUST00000167144.2
ENSMUST00000170878.2
|
Cd180
|
CD180 antigen
|
|
chr5_-_51711237
Show fit
|
0.28 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
|
|
chr1_+_171265103
Show fit
|
0.27 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor
|
|
chr10_-_79744726
Show fit
|
0.26 |
ENSMUST00000165684.8
ENSMUST00000164705.8
ENSMUST00000105378.9
ENSMUST00000170409.2
|
Med16
|
mediator complex subunit 16
|
|
chr11_-_120538928
Show fit
|
0.26 |
ENSMUST00000239158.2
ENSMUST00000026134.3
|
Myadml2
|
myeloid-associated differentiation marker-like 2
|
|
chr3_-_51248032
Show fit
|
0.26 |
ENSMUST00000062009.14
ENSMUST00000194641.6
|
Elf2
|
E74-like factor 2
|
|
chr1_+_179936757
Show fit
|
0.25 |
ENSMUST00000143176.8
ENSMUST00000135056.8
|
Cdc42bpa
|
CDC42 binding protein kinase alpha
|
|
chr8_-_62355690
Show fit
|
0.25 |
ENSMUST00000121785.9
ENSMUST00000034057.14
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr2_+_112097087
Show fit
|
0.25 |
ENSMUST00000110987.9
ENSMUST00000028549.14
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr17_+_73433422
Show fit
|
0.25 |
ENSMUST00000232703.2
|
Lclat1
|
lysocardiolipin acyltransferase 1
|
|
chr3_+_102981352
Show fit
|
0.25 |
ENSMUST00000176440.2
|
Ampd1
|
adenosine monophosphate deaminase 1
|
|
chr3_+_116356592
Show fit
|
0.24 |
ENSMUST00000029573.8
|
Lrrc39
|
leucine rich repeat containing 39
|
|
chr11_+_75546989
Show fit
|
0.24 |
ENSMUST00000136935.2
|
Myo1c
|
myosin IC
|
|
chr8_-_24928953
Show fit
|
0.24 |
ENSMUST00000052622.6
|
Tcim
|
transcriptional and immune response regulator
|
|
chr11_-_78950641
Show fit
|
0.24 |
ENSMUST00000226282.2
|
Ksr1
|
kinase suppressor of ras 1
|
|
chr1_+_63485002
Show fit
|
0.24 |
ENSMUST00000087374.10
|
Adam23
|
a disintegrin and metallopeptidase domain 23
|
|
chr2_+_27566452
Show fit
|
0.23 |
ENSMUST00000129514.8
|
Rxra
|
retinoid X receptor alpha
|
|
chr5_-_88675700
Show fit
|
0.23 |
ENSMUST00000087033.6
|
Jchain
|
immunoglobulin joining chain
|
|
chr10_+_69048506
Show fit
|
0.23 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1
|
|
chr7_-_100164007
Show fit
|
0.22 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13
|
|
chr11_-_69289052
Show fit
|
0.22 |
ENSMUST00000050140.6
|
Tmem88
|
transmembrane protein 88
|
|
chr2_-_77000936
Show fit
|
0.22 |
ENSMUST00000164114.9
ENSMUST00000049544.14
|
Ccdc141
|
coiled-coil domain containing 141
|
|
chr7_-_128020397
Show fit
|
0.22 |
ENSMUST00000147840.2
|
Rgs10
|
regulator of G-protein signalling 10
|
|
chrX_+_158086253
Show fit
|
0.21 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3
|
|
chr5_-_122140403
Show fit
|
0.21 |
ENSMUST00000134326.2
|
Cux2
|
cut-like homeobox 2
|
|
chr12_+_111387978
Show fit
|
0.20 |
ENSMUST00000222897.2
|
Exoc3l4
|
exocyst complex component 3-like 4
|
|
chr18_+_4994600
Show fit
|
0.20 |
ENSMUST00000140448.8
|
Svil
|
supervillin
|
|
chr10_+_69048914
Show fit
|
0.19 |
ENSMUST00000163497.8
ENSMUST00000164212.8
ENSMUST00000067908.14
|
Rhobtb1
|
Rho-related BTB domain containing 1
|
|
chr19_-_6065872
Show fit
|
0.19 |
ENSMUST00000164843.10
|
Capn1
|
calpain 1
|
|
chr4_+_11485945
Show fit
|
0.19 |
ENSMUST00000055372.14
ENSMUST00000059914.13
|
Virma
|
vir like m6A methyltransferase associated
|
|
chr5_-_51711204
Show fit
|
0.18 |
ENSMUST00000196968.5
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
|
|
chr15_+_102379621
Show fit
|
0.18 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2
|
|
chr3_-_123029782
Show fit
|
0.18 |
ENSMUST00000106427.8
ENSMUST00000198584.2
|
Synpo2
|
synaptopodin 2
|
|
chr1_+_131755715
Show fit
|
0.18 |
ENSMUST00000086559.7
|
Slc41a1
|
solute carrier family 41, member 1
|
|
chr11_+_75546671
Show fit
|
0.18 |
ENSMUST00000108431.3
|
Myo1c
|
myosin IC
|
|
chr5_+_64316771
Show fit
|
0.17 |
ENSMUST00000043893.13
|
Tbc1d1
|
TBC1 domain family, member 1
|
|
chr6_+_5390386
Show fit
|
0.17 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4
|
|
chr11_+_120123727
Show fit
|
0.16 |
ENSMUST00000122148.8
ENSMUST00000044985.14
|
Bahcc1
|
BAH domain and coiled-coil containing 1
|
|
chr6_+_108190163
Show fit
|
0.15 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1
|
|
chr5_-_122752508
Show fit
|
0.15 |
ENSMUST00000127220.8
ENSMUST00000031426.14
|
Ift81
|
intraflagellar transport 81
|
|
chr13_+_44994167
Show fit
|
0.14 |
ENSMUST00000173906.3
|
Jarid2
|
jumonji, AT rich interactive domain 2
|
|
chr19_-_24839219
Show fit
|
0.14 |
ENSMUST00000047666.5
|
Pgm5
|
phosphoglucomutase 5
|
|
chr1_-_88629843
Show fit
|
0.13 |
ENSMUST00000159814.2
|
Arl4c
|
ADP-ribosylation factor-like 4C
|
|
chr14_+_102078038
Show fit
|
0.13 |
ENSMUST00000159314.8
|
Lmo7
|
LIM domain only 7
|
|
chr6_+_116627635
Show fit
|
0.13 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator
|
|
chr9_-_66421868
Show fit
|
0.12 |
ENSMUST00000056890.10
|
Fbxl22
|
F-box and leucine-rich repeat protein 22
|
|
chr15_+_101152078
Show fit
|
0.12 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr16_+_29884153
Show fit
|
0.11 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1
|
|
chr18_+_89195089
Show fit
|
0.11 |
ENSMUST00000236644.2
ENSMUST00000236828.2
|
Cd226
|
CD226 antigen
|
|
chr4_+_135487016
Show fit
|
0.11 |
ENSMUST00000105854.2
|
Myom3
|
myomesin family, member 3
|
|
chr9_-_79884920
Show fit
|
0.11 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1
|