Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
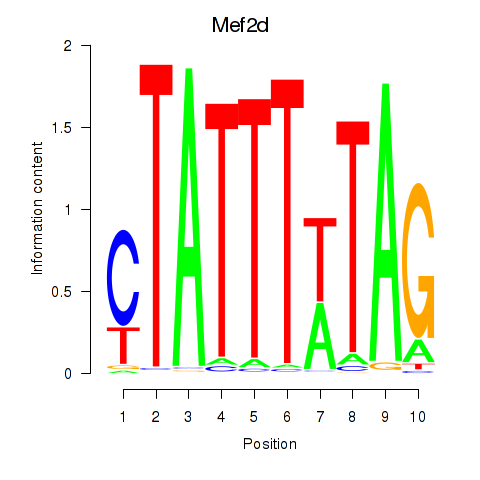
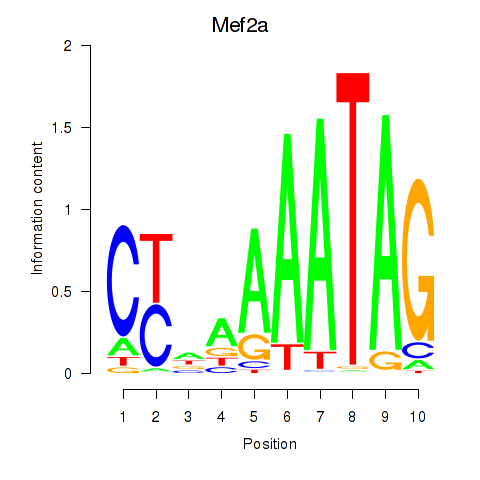
Results for Mef2d_Mef2a
Z-value: 1.65
Transcription factors associated with Mef2d_Mef2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2d
|
ENSMUSG00000001419.18 | myocyte enhancer factor 2D |
|
Mef2a
|
ENSMUSG00000030557.18 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2d | mm39_v1_chr3_+_88049633_88049790 | -0.35 | 3.6e-02 | Click! |
| Mef2a | mm39_v1_chr7_-_67022520_67022609 | -0.24 | 1.6e-01 | Click! |
Activity profile of Mef2d_Mef2a motif
Sorted Z-values of Mef2d_Mef2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_97066937 | 11.51 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr14_+_55813074 | 11.32 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr11_+_98274637 | 8.28 |
ENSMUST00000008021.3
|
Tcap
|
titin-cap |
| chr1_-_66974492 | 8.07 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr1_-_66974694 | 7.51 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr5_+_122239007 | 6.93 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr5_+_122239030 | 6.59 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr19_+_20579322 | 6.15 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr7_-_48497771 | 6.10 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr7_+_141995545 | 5.91 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr10_+_87694117 | 5.78 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr14_-_52151537 | 5.55 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr3_-_122828592 | 5.46 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr14_-_52151026 | 5.35 |
ENSMUST00000228164.2
|
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr5_+_135916764 | 4.93 |
ENSMUST00000005077.7
|
Hspb1
|
heat shock protein 1 |
| chr8_+_110717062 | 4.71 |
ENSMUST00000001720.14
ENSMUST00000143741.2 |
Tat
|
tyrosine aminotransferase |
| chr5_+_135916847 | 4.60 |
ENSMUST00000111155.2
|
Hspb1
|
heat shock protein 1 |
| chr9_+_74860133 | 4.58 |
ENSMUST00000215370.2
|
Fam214a
|
family with sequence similarity 214, member A |
| chr14_-_34310602 | 4.45 |
ENSMUST00000064098.14
ENSMUST00000090040.12 ENSMUST00000022330.9 ENSMUST00000022327.13 |
Ldb3
|
LIM domain binding 3 |
| chr14_-_34310637 | 4.43 |
ENSMUST00000227819.2
|
Ldb3
|
LIM domain binding 3 |
| chr1_-_66984178 | 4.36 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr11_+_67167950 | 4.33 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr9_-_119852624 | 4.17 |
ENSMUST00000111635.4
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr17_+_85335775 | 4.05 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr4_-_134099840 | 3.86 |
ENSMUST00000030643.3
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr10_-_88440869 | 3.86 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr15_+_25940912 | 3.79 |
ENSMUST00000226438.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr1_-_172047282 | 3.74 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chr1_-_66984521 | 3.72 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chr15_+_25940781 | 3.68 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr6_-_71239216 | 3.53 |
ENSMUST00000129630.3
ENSMUST00000114186.9 ENSMUST00000074301.10 |
Smyd1
|
SET and MYND domain containing 1 |
| chr11_+_67689094 | 3.51 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr8_+_15107646 | 3.50 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr13_-_24118139 | 3.33 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr7_+_141996067 | 3.33 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr7_+_142052569 | 3.28 |
ENSMUST00000078497.15
ENSMUST00000105953.10 ENSMUST00000179658.8 ENSMUST00000105954.10 ENSMUST00000105952.10 ENSMUST00000105955.8 ENSMUST00000074187.13 ENSMUST00000169299.9 ENSMUST00000105957.10 ENSMUST00000180152.8 ENSMUST00000105950.11 ENSMUST00000105958.10 ENSMUST00000105949.8 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr15_+_25940931 | 3.25 |
ENSMUST00000110438.3
|
Retreg1
|
reticulophagy regulator 1 |
| chr2_-_25359752 | 3.23 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr7_+_80707328 | 3.15 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chr11_+_67090878 | 2.95 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr2_+_90948481 | 2.91 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr14_+_66205932 | 2.89 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr3_+_102981326 | 2.84 |
ENSMUST00000090715.13
ENSMUST00000155034.6 |
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr1_+_40844739 | 2.83 |
ENSMUST00000114765.4
|
Tmem182
|
transmembrane protein 182 |
| chr11_-_94867153 | 2.78 |
ENSMUST00000103162.8
ENSMUST00000166320.8 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chrX_+_100492684 | 2.73 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chrX_-_94521712 | 2.70 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr9_-_31824758 | 2.64 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chr2_+_102488985 | 2.63 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_-_127805518 | 2.62 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr10_-_88440996 | 2.55 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chrX_+_156481906 | 2.55 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr10_-_108846816 | 2.49 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr14_-_34310438 | 2.49 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr4_+_133280680 | 2.39 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr13_+_89687915 | 2.26 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr2_-_25360043 | 2.22 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr2_-_33261498 | 2.20 |
ENSMUST00000113165.8
|
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chrX_+_156482116 | 2.18 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr13_+_24118417 | 2.15 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr1_+_182392559 | 2.12 |
ENSMUST00000168514.7
|
Capn8
|
calpain 8 |
| chr9_+_121606750 | 2.12 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40 |
| chr2_-_33261411 | 2.08 |
ENSMUST00000131298.7
ENSMUST00000091039.5 ENSMUST00000042615.13 |
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr17_-_74257164 | 2.07 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr7_+_127603083 | 2.05 |
ENSMUST00000106248.8
|
Trim72
|
tripartite motif-containing 72 |
| chr3_+_101993787 | 2.04 |
ENSMUST00000165540.9
ENSMUST00000164123.2 |
Casq2
|
calsequestrin 2 |
| chr2_+_69500444 | 2.00 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr3_+_101993731 | 1.97 |
ENSMUST00000029454.12
|
Casq2
|
calsequestrin 2 |
| chr14_-_20718337 | 1.97 |
ENSMUST00000057090.12
ENSMUST00000117386.2 |
Synpo2l
|
synaptopodin 2-like |
| chr18_-_3281089 | 1.97 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr3_-_142101339 | 1.96 |
ENSMUST00000198381.5
ENSMUST00000090134.12 ENSMUST00000196908.5 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr9_+_110592709 | 1.94 |
ENSMUST00000079784.12
|
Myl3
|
myosin, light polypeptide 3 |
| chr13_-_36918424 | 1.93 |
ENSMUST00000037623.15
|
Nrn1
|
neuritin 1 |
| chr13_+_23868175 | 1.93 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr9_-_70048766 | 1.91 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr18_+_67597929 | 1.91 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chr3_-_95811993 | 1.91 |
ENSMUST00000147962.3
ENSMUST00000036181.15 |
Car14
|
carbonic anhydrase 14 |
| chr5_-_113044216 | 1.90 |
ENSMUST00000086617.11
|
Myo18b
|
myosin XVIIIb |
| chr17_-_48739874 | 1.90 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr19_-_56378309 | 1.89 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr2_-_32584132 | 1.88 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr6_+_121277186 | 1.84 |
ENSMUST00000064580.14
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr11_+_70548513 | 1.83 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr2_+_71884943 | 1.83 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr6_+_121277693 | 1.83 |
ENSMUST00000142419.2
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr18_-_39062201 | 1.82 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr19_-_56378459 | 1.81 |
ENSMUST00000040711.15
ENSMUST00000095947.11 ENSMUST00000073536.13 |
Nrap
|
nebulin-related anchoring protein |
| chr7_+_101750943 | 1.80 |
ENSMUST00000033300.4
|
Art1
|
ADP-ribosyltransferase 1 |
| chr1_+_132996237 | 1.80 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chrX_-_8059597 | 1.77 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr7_-_30754792 | 1.77 |
ENSMUST00000206328.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_30755007 | 1.71 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_4517559 | 1.70 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr12_+_41074089 | 1.65 |
ENSMUST00000132121.8
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_41074328 | 1.64 |
ENSMUST00000134965.8
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr15_-_78090591 | 1.61 |
ENSMUST00000120592.2
|
Pvalb
|
parvalbumin |
| chr1_+_182392577 | 1.61 |
ENSMUST00000048941.14
|
Capn8
|
calpain 8 |
| chr8_+_46111703 | 1.59 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr18_-_39062514 | 1.59 |
ENSMUST00000235922.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr11_+_94219046 | 1.57 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr2_+_32515442 | 1.56 |
ENSMUST00000113277.8
ENSMUST00000195721.6 |
Ak1
|
adenylate kinase 1 |
| chr11_+_70548022 | 1.55 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr5_-_113229445 | 1.53 |
ENSMUST00000131708.2
ENSMUST00000117143.8 ENSMUST00000119627.8 |
Crybb3
|
crystallin, beta B3 |
| chr13_+_102830104 | 1.53 |
ENSMUST00000172138.2
|
Cd180
|
CD180 antigen |
| chr10_-_24588030 | 1.49 |
ENSMUST00000105520.8
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr1_-_172125555 | 1.46 |
ENSMUST00000085913.11
ENSMUST00000097464.4 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr7_-_4517608 | 1.46 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr12_+_108300599 | 1.45 |
ENSMUST00000021684.6
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr3_-_142101418 | 1.44 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr11_-_77784922 | 1.39 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr17_-_45903494 | 1.38 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr15_+_25940859 | 1.37 |
ENSMUST00000226750.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr15_-_76906832 | 1.36 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr10_-_24587884 | 1.36 |
ENSMUST00000135846.2
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr16_+_8331293 | 1.35 |
ENSMUST00000065987.14
ENSMUST00000115838.8 ENSMUST00000115839.9 |
Abat
|
4-aminobutyrate aminotransferase |
| chr16_-_35589726 | 1.34 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr3_-_95789505 | 1.33 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr3_-_137849113 | 1.32 |
ENSMUST00000098580.6
|
Mttp
|
microsomal triglyceride transfer protein |
| chr6_+_53264255 | 1.30 |
ENSMUST00000203528.3
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr14_-_20714634 | 1.29 |
ENSMUST00000119483.2
|
Synpo2l
|
synaptopodin 2-like |
| chr5_+_92534738 | 1.29 |
ENSMUST00000128246.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr12_-_84447702 | 1.29 |
ENSMUST00000122194.8
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr19_-_34856853 | 1.26 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chrX_+_106299484 | 1.21 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr1_+_180678677 | 1.20 |
ENSMUST00000038091.8
|
Sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr17_-_36208265 | 1.20 |
ENSMUST00000148721.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr17_+_50816296 | 1.20 |
ENSMUST00000043938.8
|
Plcl2
|
phospholipase C-like 2 |
| chr2_-_77000878 | 1.19 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr18_-_12995395 | 1.18 |
ENSMUST00000121888.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr6_-_116050081 | 1.18 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr18_-_12995261 | 1.17 |
ENSMUST00000234427.2
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr3_+_102981352 | 1.17 |
ENSMUST00000176440.2
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr8_+_46111310 | 1.16 |
ENSMUST00000153798.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr9_+_102382949 | 1.15 |
ENSMUST00000039390.6
|
Ky
|
kyphoscoliosis peptidase |
| chr11_-_69260203 | 1.13 |
ENSMUST00000092971.13
ENSMUST00000108661.8 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr3_+_96484294 | 1.13 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chr2_+_3514071 | 1.13 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr11_+_77353431 | 1.11 |
ENSMUST00000130255.2
|
Coro6
|
coronin 6 |
| chr8_+_77628916 | 1.07 |
ENSMUST00000109912.8
ENSMUST00000128862.2 ENSMUST00000109911.8 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr11_+_108811626 | 1.05 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr12_-_84447625 | 1.05 |
ENSMUST00000117286.2
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr15_+_9140685 | 1.04 |
ENSMUST00000090380.6
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr16_-_4340920 | 1.04 |
ENSMUST00000090500.10
ENSMUST00000023161.8 |
Srl
|
sarcalumenin |
| chr7_+_27879650 | 1.03 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr5_-_24745970 | 1.03 |
ENSMUST00000117900.8
|
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr18_-_3280999 | 1.03 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr4_-_73869071 | 1.02 |
ENSMUST00000095023.2
ENSMUST00000030101.4 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr1_+_135727571 | 1.02 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr8_+_46111361 | 1.02 |
ENSMUST00000210946.2
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_+_131671751 | 1.00 |
ENSMUST00000049027.10
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr4_-_44066960 | 1.00 |
ENSMUST00000173234.8
ENSMUST00000173274.2 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr6_+_42263609 | 0.99 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr2_+_32518402 | 0.98 |
ENSMUST00000156578.8
|
Ak1
|
adenylate kinase 1 |
| chr2_+_19662529 | 0.98 |
ENSMUST00000052168.6
|
Otud1
|
OTU domain containing 1 |
| chr4_+_134042423 | 0.97 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr16_-_90866032 | 0.97 |
ENSMUST00000035689.8
ENSMUST00000114076.2 |
4932438H23Rik
|
RIKEN cDNA 4932438H23 gene |
| chr1_+_16758731 | 0.97 |
ENSMUST00000190366.2
|
Ly96
|
lymphocyte antigen 96 |
| chr7_-_101749433 | 0.95 |
ENSMUST00000106937.8
|
Art5
|
ADP-ribosyltransferase 5 |
| chr1_+_16758629 | 0.95 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr3_+_32871669 | 0.95 |
ENSMUST00000072312.12
ENSMUST00000108228.8 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr17_-_45903188 | 0.94 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr5_+_28370687 | 0.94 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chr7_+_27878894 | 0.94 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr18_+_37606591 | 0.93 |
ENSMUST00000050034.3
|
Pcdhb15
|
protocadherin beta 15 |
| chr1_+_135727140 | 0.93 |
ENSMUST00000152208.8
ENSMUST00000152075.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr1_-_64161415 | 0.90 |
ENSMUST00000135075.2
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr7_-_4517367 | 0.90 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr1_+_187341225 | 0.90 |
ENSMUST00000110939.8
|
Esrrg
|
estrogen-related receptor gamma |
| chr2_+_27055245 | 0.89 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr1_+_135727228 | 0.89 |
ENSMUST00000154463.8
ENSMUST00000139986.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chrX_+_163221035 | 0.87 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr17_-_45903410 | 0.86 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr15_-_95426108 | 0.86 |
ENSMUST00000075275.3
|
Nell2
|
NEL-like 2 |
| chr9_-_58462720 | 0.86 |
ENSMUST00000165365.3
|
Cd276
|
CD276 antigen |
| chr11_+_94218810 | 0.85 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr1_+_130947580 | 0.85 |
ENSMUST00000016673.6
|
Il10
|
interleukin 10 |
| chr7_-_127423641 | 0.85 |
ENSMUST00000106267.5
|
Stx1b
|
syntaxin 1B |
| chr15_-_98705791 | 0.84 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr1_-_89942299 | 0.83 |
ENSMUST00000086882.8
ENSMUST00000097656.10 |
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr2_-_48839218 | 0.83 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr5_-_51711237 | 0.83 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr10_+_34359395 | 0.83 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr3_+_137849189 | 0.82 |
ENSMUST00000040321.13
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr3_-_123029782 | 0.82 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chrX_-_46981273 | 0.81 |
ENSMUST00000153548.9
ENSMUST00000141084.3 |
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr6_+_95094721 | 0.80 |
ENSMUST00000032107.10
ENSMUST00000119582.3 |
Kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr6_+_137387718 | 0.79 |
ENSMUST00000167002.4
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_187340952 | 0.79 |
ENSMUST00000127489.8
|
Esrrg
|
estrogen-related receptor gamma |
| chr10_+_69621337 | 0.78 |
ENSMUST00000182993.8
|
Ank3
|
ankyrin 3, epithelial |
| chr6_+_42263644 | 0.77 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr9_-_110571645 | 0.77 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr3_+_122277372 | 0.77 |
ENSMUST00000197073.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr10_-_107330580 | 0.76 |
ENSMUST00000044210.5
|
Myf6
|
myogenic factor 6 |
| chr19_+_38253077 | 0.75 |
ENSMUST00000198045.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr5_+_138278502 | 0.75 |
ENSMUST00000160729.8
|
Stag3
|
stromal antigen 3 |
| chr14_+_66074751 | 0.75 |
ENSMUST00000022614.7
|
Ccdc25
|
coiled-coil domain containing 25 |
| chr7_+_16609227 | 0.74 |
ENSMUST00000108493.3
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr5_-_25047577 | 0.73 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr16_-_43836681 | 0.72 |
ENSMUST00000036174.10
|
Gramd1c
|
GRAM domain containing 1C |
| chr3_+_20011251 | 0.72 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2d_Mef2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 2.0 | 6.1 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 1.8 | 10.9 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.4 | 8.6 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 1.4 | 11.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.3 | 12.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.1 | 3.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 1.1 | 9.5 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.9 | 3.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.9 | 5.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.8 | 5.8 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.7 | 6.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.7 | 4.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.6 | 1.9 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.6 | 2.2 | GO:0007522 | visceral muscle development(GO:0007522) |
| 0.5 | 4.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 1.6 | GO:2000184 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.5 | 3.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.5 | 1.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.5 | 2.9 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.5 | 4.2 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.5 | 1.4 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.5 | 3.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.5 | 1.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.4 | 4.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.4 | 4.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 | 19.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.4 | 2.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.4 | 2.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.4 | 4.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 3.5 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.4 | 1.9 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.4 | 2.6 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 1.5 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 1.8 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 1.5 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.3 | 1.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.3 | 12.7 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.3 | 25.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.3 | 2.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 1.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.3 | 0.9 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.3 | 2.9 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.3 | 0.9 | GO:0060300 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) |
| 0.3 | 1.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 1.6 | GO:0061181 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.3 | 1.1 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.3 | 2.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.3 | 3.1 | GO:0042637 | catagen(GO:0042637) |
| 0.3 | 2.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 0.9 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.2 | 0.6 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 2.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 0.6 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.2 | 1.0 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 | 1.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 0.4 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 0.5 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.2 | 0.9 | GO:1904048 | negative regulation of synaptic vesicle recycling(GO:1903422) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.2 | 0.5 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 2.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 1.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 1.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 1.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.5 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.1 | 0.5 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 0.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 24.1 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.9 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.6 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.5 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 2.3 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 | 1.5 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 1.0 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 3.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.5 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 1.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.4 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 4.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 1.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.7 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.6 | GO:1900042 | interleukin-4 secretion(GO:0072602) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 1.0 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 1.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.9 | GO:0070050 | development of secondary female sexual characteristics(GO:0046543) neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 1.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 3.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 1.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 3.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 1.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.7 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.8 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 3.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 3.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.9 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 1.0 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.7 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.9 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 2.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 2.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.8 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 1.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.6 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 1.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.3 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 1.9 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 3.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 1.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 3.5 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 1.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 1.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0097106 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) postsynaptic density organization(GO:0097106) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.5 | 19.4 | GO:0005861 | troponin complex(GO:0005861) |
| 1.3 | 25.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.2 | 3.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 1.1 | 3.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 1.1 | 8.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.6 | 15.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.6 | 5.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 1.6 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.5 | 5.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.4 | 2.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.4 | 1.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 1.0 | GO:0090537 | CERF complex(GO:0090537) |
| 0.3 | 2.0 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 2.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 0.9 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.3 | 3.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 4.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 18.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 1.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.7 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.2 | 1.0 | GO:0000802 | transverse filament(GO:0000802) |
| 0.2 | 2.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 2.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 2.1 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 0.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 12.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 3.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 28.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.5 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 1.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 4.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 9.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 11.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.6 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 4.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 3.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 2.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.3 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0032590 | dendrite membrane(GO:0032590) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.6 | 4.7 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.5 | 6.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.4 | 5.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.3 | 13.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.3 | 3.9 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 1.2 | 6.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.0 | 28.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.8 | 2.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.8 | 5.5 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.6 | 1.9 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.6 | 3.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.6 | 2.9 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.5 | 3.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.5 | 2.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.4 | 2.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 2.1 | GO:0030151 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.4 | 4.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 1.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 1.0 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.3 | 1.9 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 1.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.3 | 15.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 0.9 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.3 | 4.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 3.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.8 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 2.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 9.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 1.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 1.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 1.0 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.2 | 4.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 0.5 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 2.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.7 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 3.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 1.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 3.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 3.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 5.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 2.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.8 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.8 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 3.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 2.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 1.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 3.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 2.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 3.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.6 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 27.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 2.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 2.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0015491 | cation:cation antiporter activity(GO:0015491) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 3.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.3 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 1.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 10.8 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 9.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 3.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 66.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 6.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 2.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 3.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 4.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 9.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.8 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 2.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 3.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 4.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 5.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.0 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |